I don't think this will become a priority in the near future - if anything, this should go into a separate package. Closing for now.
Closed grst closed 9 months ago
I don't think this will become a priority in the near future - if anything, this should go into a separate package. Closing for now.
@jfx319 wrote:
At least several factors affect observed categories:
Adding to the sources you've already cited, this illustration from Dupic et al 2019 emphasizes the different a priori expectation of the beta vs alpha chain:
Separate from the allelic exclusion bias, this figure from Redmond et al 2016 illustrates the 2 fold expression bias between alpha and beta: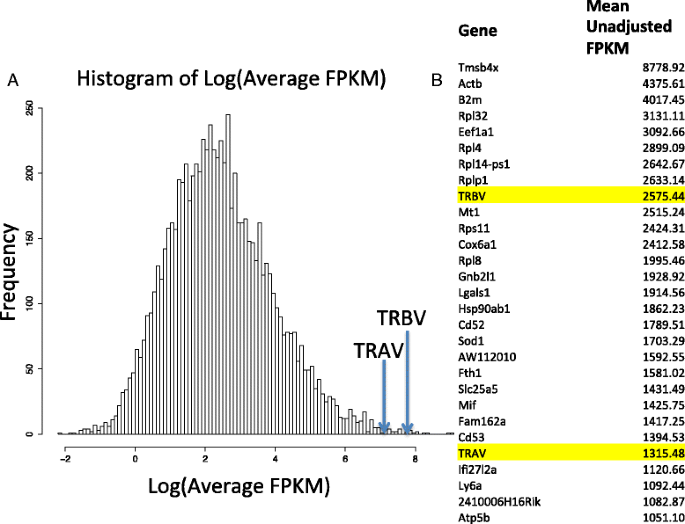
These biological mechanisms contribute the the observed frequencies. But what is measured is additionally confounded by signal dropout and by doublets.
Single-cell considerations: