It's solved. when I used harmony_integrate function of scanpy, it's worked.
sce.pp.harmony_integrate(adata, 'aligned_dataset',basis='X_uce')Closed zhou-1314 closed 10 months ago
It's solved. when I used harmony_integrate function of scanpy, it's worked.
sce.pp.harmony_integrate(adata, 'aligned_dataset',basis='X_uce')Thanks for sharing your experience.
If we search the web for "precompiled NUM_THREADS exceeded" we can find many results with suggestions for how to solve this issue.
For example, this Stackoverflow post suggests that we can set the NUM_THREADS option like this:
os.environ['OPENBLAS_NUM_THREADS'] = '1'and then the sklearn.KMeans function should run without error.
I do not know whether or not this will fix your issue, because I don't know your system environment (python version, package versions, etc.).
First of all, thank you very much for migrating the Harmony algorithm from R to Python. Today, I encountered the following error while integrating 318,720 cells from different datasets using harmonypy.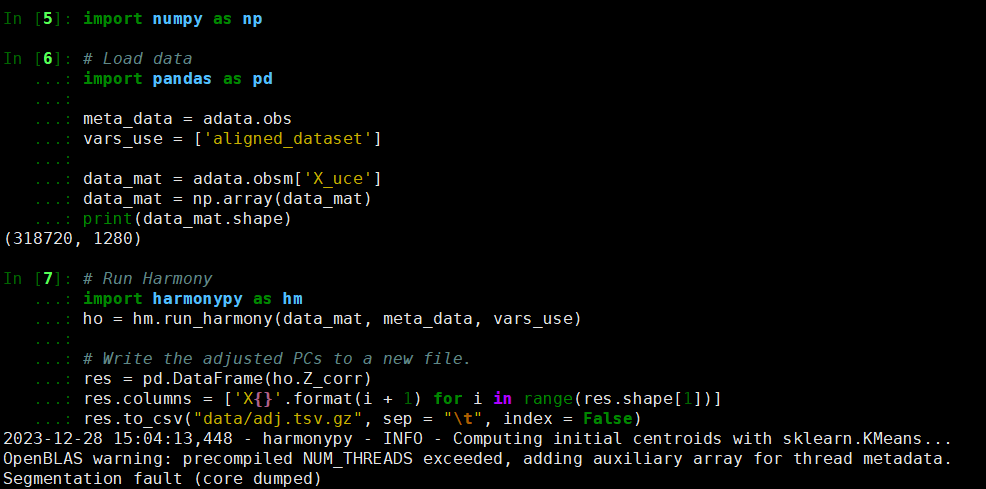 Hope to receive your help. Thank you.
Hope to receive your help. Thank you.