Can you please re-install the package and try again?
Closed drake77777 closed 2 months ago
Can you please re-install the package and try again?
Thank you for your kindly help, but that dindn't work, tried reinstall hdWGCNA and Seurat package, also didn't work, now trying reinstall dependent packages.
Thanks a lot! After i reinstall hdWGCNA three times, it worked!
Hello! This package is excellect. I ran it perfectly first time when i loaded a seurat v4 template object from my other project, but when I changed to Seurat v5 object, it started to show this error and even that seurat v4 object encountered this error too. i could't find where went wrong, would you please help me with this? I tried to convert a v5 assay to a v4 assay, still had the same error.
package version : hdWGCNA_0.3.00 Seurat_5.0.2 .
Here is the error message when i useing this function-ModuleEigengenes :
Here are the seurat v5 and v4 objects , v5 :An object of class Seurat
23825 features across 22096 samples within 1 assay Active assay: RNA (23825 features, 4000 variable features) 3 layers present: scale.data, data, counts 4 dimensional reductions calculated: pca, integrated.Harmony, umap, tsne
v4 : An object of class Seurat
28830 features across 19468 samples within 2 assays Active assay: RNA (26830 features, 0 variable features) 2 layers present: counts, data 1 other assay present: integrated 2 dimensional reductions calculated: pca, umap
This is my code :
R session info
sessionInfo() R version 4.3.1 (2023-06-16 ucrt) Platform: x86_64-w64-mingw32/x64 (64-bit) Running under: Windows 11 x64 (build 22621)
Matrix products: default
locale: [1] LC_COLLATE=Chinese (Simplified)_China.utf8 LC_CTYPE=Chinese (Simplified)_China.utf8 LC_MONETARY=Chinese (Simplified)_China.utf8 [4] LC_NUMERIC=C LC_TIME=Chinese (Simplified)_China.utf8
time zone: Etc/GMT-8 tzcode source: internal
attached base packages: [1] stats graphics grDevices utils datasets methods base
other attached packages: [1] patchwork_1.2.0 cowplot_1.1.3 lubridate_1.9.3 forcats_1.0.0 stringr_1.5.1
[6] dplyr_1.1.4 purrr_1.0.2 readr_2.1.5 tidyr_1.3.0 tibble_3.2.1
[11] tidyverse_2.0.0 hdWGCNA_0.3.00 igraph_1.6.0 WGCNA_1.72-5 fastcluster_1.2.6
[16] dynamicTreeCut_1.63-1 ggrepel_0.9.5 ggplot2_3.5.0 Seurat_5.0.2 SeuratObject_5.0.1
[21] sp_2.1-2 harmony_1.2.0 Rcpp_1.0.11
loaded via a namespace (and not attached): [1] IRanges_2.34.1 R.methodsS3_1.8.2 urlchecker_1.0.1 nnet_7.3-19 ggfittext_0.10.2
[6] goftest_1.2-3 phytools_2.1-1 Biostrings_2.68.1 vctrs_0.6.4 spatstat.random_3.2-2
[11] digest_0.6.33 png_0.1-8 shape_1.4.6.1 proxy_0.4-27 deldir_2.0-2
[16] parallelly_1.37.0 combinat_0.0-8 MASS_7.3-60 reshape2_1.4.4 httpuv_1.6.11
[21] foreach_1.5.2 BiocGenerics_0.46.0 qvalue_2.32.0 withr_3.0.0 ggrastr_1.0.2
[26] xfun_0.42 ggfun_0.1.4 ellipsis_0.3.2 survival_3.5-8 memoise_2.0.1
[31] ggbeeswarm_0.7.2 clusterProfiler_4.8.3 gson_0.1.0 profvis_0.3.8 tidytree_0.4.6
[36] zoo_1.8-12 GlobalOptions_0.1.2 pbapply_1.7-2 R.oo_1.26.0 Formula_1.2-5
[41] rematch2_2.1.2 KEGGREST_1.40.1 promises_1.2.1 scatterplot3d_0.3-44 httr_1.4.7
[46] downloader_0.4 globals_0.16.2 fitdistrplus_1.1-11 rstudioapi_0.15.0 miniUI_0.1.1.1
[51] generics_0.1.3 DOSE_3.26.2 base64enc_0.1-3 curl_5.2.0 S4Vectors_0.38.2
[56] zlibbioc_1.46.0 ggraph_2.1.0 polyclip_1.10-6 GenomeInfoDbData_1.2.11 quadprog_1.5-8
[61] xtable_1.8-4 doParallel_1.0.17 evaluate_0.23 preprocessCore_1.64.0 hms_1.1.3
[66] irlba_2.3.5.1 colorspace_2.1-0 ROCR_1.0-11 reticulate_1.34.0 spatstat.data_3.0-4
[71] magrittr_2.0.3 lmtest_0.9-40 later_1.3.1 viridis_0.6.5 ggtree_3.8.2
[76] lattice_0.21-8 spatstat.geom_3.2-7 future.apply_1.11.1 scattermore_1.2 shadowtext_0.1.3
[81] matrixStats_1.2.0 RcppAnnoy_0.0.21 Hmisc_5.1-1 pillar_1.9.0 nlme_3.1-162
[86] iterators_1.0.14 compiler_4.3.1 RSpectra_0.16-1 stringi_1.7.12 tensor_1.5
[91] devtools_2.4.5 plyr_1.8.9 crayon_1.5.2 abind_1.4-5 gridGraphics_0.5-1
[96] graphlayouts_1.0.2 bit_4.0.5 fastmatch_1.1-4 codetools_0.2-19 paletteer_1.6.0
[101] GetoptLong_1.0.5 plotly_4.10.4 mime_0.12 splines_4.3.1 circlize_0.4.16
[106] fastDummies_1.7.3 HDO.db_0.99.1 knitr_1.45 blob_1.2.4 utf8_1.2.3
[111] clue_0.3-65 fs_1.6.3 listenv_0.9.1 checkmate_2.3.1 pkgbuild_1.4.3
[116] expm_0.999-9 ggplotify_0.1.2 Matrix_1.6-4 tzdb_0.4.0 tweenr_2.0.2
[121] pkgconfig_2.0.3 pheatmap_1.0.12 tools_4.3.1 cachem_1.0.8 RSQLite_2.3.5
[126] viridisLite_0.4.2 DBI_1.2.2 numDeriv_2016.8-1.1 impute_1.76.0 fastmap_1.1.1
[131] rmarkdown_2.25 scales_1.3.0 grid_4.3.1 usethis_2.2.3 ica_1.0-3
[136] FNN_1.1.4 coda_0.19-4.1 BiocManager_1.30.22 dotCall64_1.1-1 RANN_2.6.1
[141] rpart_4.1.19 farver_2.1.1 tidygraph_1.3.0 scatterpie_0.2.1 deeptime_1.0.1
[146] foreign_0.8-84 cli_3.6.1 stats4_4.3.1 tester_0.1.7 leiden_0.4.3.1
[151] lifecycle_1.0.4 uwot_0.1.16 Biobase_2.60.0 sessioninfo_1.2.2 backports_1.4.1
[156] BiocParallel_1.34.2 timechange_0.3.0 gtable_0.3.4 rjson_0.2.21 ggridges_0.5.6
[161] progressr_0.14.0 parallel_4.3.1 ape_5.7-1 jsonlite_1.8.8 RcppHNSW_0.5.0
[166] bitops_1.0-7 bit64_4.0.5 Rtsne_0.17 yulab.utils_0.1.4 spatstat.utils_3.0-4
[171] GOSemSim_2.26.1 R.utils_2.12.3 lazyeval_0.2.2 shiny_1.8.0 htmltools_0.5.7
[176] enrichplot_1.20.3 GO.db_3.17.0 sctransform_0.4.1 glue_1.6.2 spam_2.10-0
[181] XVector_0.40.0 RCurl_1.98-1.13 treeio_1.24.3 mnormt_2.1.1 gridExtra_2.3
[186] R6_2.5.1 labeling_0.4.3 cluster_2.1.4 pkgload_1.3.4 aplot_0.2.2
[191] GenomeInfoDb_1.36.3 tidyselect_1.2.0 vipor_0.4.7 htmlTable_2.4.2 maps_3.4.2
[196] ggforce_0.4.1 AnnotationDbi_1.62.2 future_1.33.1 munsell_0.5.0 KernSmooth_2.23-22
[201] optimParallel_1.0-2 data.table_1.14.10 htmlwidgets_1.6.4 fgsea_1.26.0 ComplexHeatmap_2.16.0
[206] RColorBrewer_1.1-3 rlang_1.1.1 clusterGeneration_1.3.8 spatstat.sparse_3.0-3 spatstat.explore_3.2-5 [211] remotes_2.4.2.1 ggnewscale_0.4.10 phangorn_2.11.1 fansi_1.0.5 beeswarm_0.4.0
Screenshots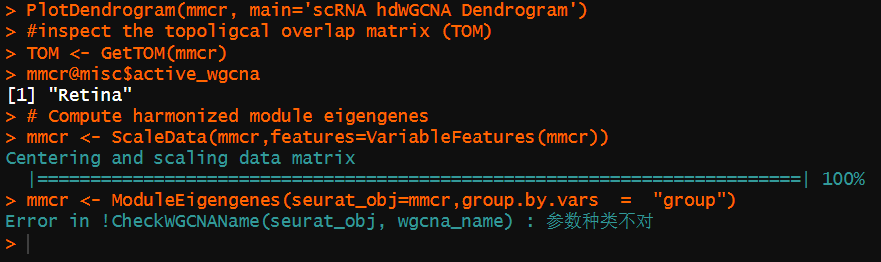
More than one problem? If you have more than one problem with hdWGCNA, create separate issues for your individual problems.