Hello,
Are you able to reproduce this error on the tutorial dataset? I tried and I am not able to. It is very difficult for me to help resolve issues unless I can reproduce the error on my side.
Open liuxiawei opened 1 month ago
Hello,
Are you able to reproduce this error on the tutorial dataset? I tried and I am not able to. It is very difficult for me to help resolve issues unless I can reproduce the error on my side.
I'm very sad that the demo code is running right way. This is my seurat object. Could you please help to check it ?
str(ta_obj)
Formal class 'Seurat' [package "SeuratObject"] with 13 slots
..@ assays :List of 2
.. ..$ Spatial:Formal class 'Assay' [package "SeuratObject"] with 8 slots
.. .. .. ..@ counts :Formal class 'dgCMatrix' [package "Matrix"] with 6 slots
.. .. .. .. .. ..@ i : int [1:469954] 350 2731 3006 4777 8380 8959 10973 12171 13098 14525 ...
.. .. .. .. .. ..@ p : int [1:492] 0 21 605 2046 2601 2931 4209 6214 7711 7774 ...
.. .. .. .. .. ..@ Dim : int [1:2] 20092 491
.. .. .. .. .. ..@ Dimnames:List of 2
.. .. .. .. .. .. ..$ : chr [1:20092] "Gene0000010" "Gene0000020" "Gene0000030" "Gene0000040" ...
.. .. .. .. .. .. ..$ : chr [1:491] "36077725294100" "36077725294200" "36077725294300" "36077725294400" ...
.. .. .. .. .. ..@ x : num [1:469954] 1 1 1 1 1 1 1 2 1 4 ...
.. .. .. .. .. ..@ factors : list()
.. .. .. ..@ data :Formal class 'dgCMatrix' [package "Matrix"] with 6 slots
.. .. .. .. .. ..@ i : int [1:469954] 350 2731 3006 4777 8380 8959 10973 12171 13098 14525 ...
.. .. .. .. .. ..@ p : int [1:492] 0 21 605 2046 2601 2931 4209 6214 7711 7774 ...
.. .. .. .. .. ..@ Dim : int [1:2] 20092 491
.. .. .. .. .. ..@ Dimnames:List of 2
.. .. .. .. .. .. ..$ : chr [1:20092] "Gene0000010" "Gene0000020" "Gene0000030" "Gene0000040" ...
.. .. .. .. .. .. ..$ : chr [1:491] "36077725294100" "36077725294200" "36077725294300" "36077725294400" ...
.. .. .. .. .. ..@ x : num [1:469954] 5.75 5.75 5.75 5.75 5.75 ...
.. .. .. .. .. ..@ factors : list()
.. .. .. ..@ scale.data : num[0 , 0 ]
.. .. .. ..@ assay.orig : NULL
.. .. .. ..@ var.features : chr(0)
.. .. .. ..@ meta.features:'data.frame': 20092 obs. of 3 variables:
.. .. .. .. ..$ n_cells : int [1:20092] 55 22 55 222 99 26 50 157 106 15 ...
.. .. .. .. ..$ n_counts: int [1:20092] 92 36 86 415 167 49 73 280 171 21 ...
.. .. .. .. ..$ mean_umi: num [1:20092] 1.67 1.64 1.56 1.87 1.69 ...
.. .. .. ..@ misc : Named list()
.. .. .. ..@ key : chr "spatial_"
.. ..$ SCT :Formal class 'SCTAssay' [package "Seurat"] with 9 slots
.. .. .. ..@ SCTModel.list:List of 1
.. .. .. .. ..$ model1:Formal class 'SCTModel' [package "Seurat"] with 7 slots
.. .. .. .. .. .. ..@ feature.attributes:'data.frame': 12084 obs. of 12 variables:
.. .. .. .. .. .. .. ..$ detection_rate : num [1:12084] 0.053 0.0244 0.0489 0.2424 0.0998 ...
.. .. .. .. .. .. .. ..$ gmean : num [1:12084] 0.0538 0.0239 0.0428 0.2671 0.1037 ...
.. .. .. .. .. .. .. ..$ variance : num [1:12084] 0.288 0.09 0.132 0.925 0.421 ...
.. .. .. .. .. .. .. ..$ residual_mean : num [1:12084] 0.01555 -0.00331 -0.00583 0.00322 -0.00609 ...
.. .. .. .. .. .. .. ..$ residual_variance : num [1:12084] 1.125 0.411 0.603 1.014 0.827 ...
.. .. .. .. .. .. .. ..$ theta : num [1:12084] 0.0847 0.0384 0.0681 0.337 0.1509 ...
.. .. .. .. .. .. .. ..$ (Intercept) : num [1:12084] -10.86 -11.67 -11.09 -9.26 -10.19 ...
.. .. .. .. .. .. .. ..$ log_umi : num [1:12084] 2.3 2.3 2.3 2.3 2.3 ...
.. .. .. .. .. .. .. ..$ genes_log_gmean_step1: logi [1:12084] FALSE TRUE FALSE FALSE FALSE FALSE ...
.. .. .. .. .. .. .. ..$ step1_theta : num [1:12084] NA 0.0393 NA NA NA ...
.. .. .. .. .. .. .. ..$ step1_(Intercept) : num [1:12084] NA -11.8 NA NA NA ...
.. .. .. .. .. .. .. ..$ step1_log_umi : num [1:12084] NA 2.3 NA NA NA ...
.. .. .. .. .. .. ..@ cell.attributes :'data.frame': 491 obs. of 3 variables:
.. .. .. .. .. .. .. ..$ umi : num [1:491] 32 2966 7874 2277 1121 ...
.. .. .. .. .. .. .. ..$ log_umi : num [1:491] 1.51 3.47 3.9 3.36 3.05 ...
.. .. .. .. .. .. .. ..$ cells_step1: logi [1:491] TRUE TRUE TRUE TRUE TRUE TRUE ...
.. .. .. .. .. .. ..@ clips :List of 2
.. .. .. .. .. .. .. ..$ vst: num [1:2] -22.2 22.2
.. .. .. .. .. .. .. ..$ sct: num [1:2] -4.05 4.05
.. .. .. .. .. .. ..@ umi.assay : chr "Spatial"
.. .. .. .. .. .. ..@ model : chr "y ~ log_umi"
.. .. .. .. .. .. ..@ arguments :List of 33
.. .. .. .. .. .. .. ..$ glmGamPoi_check : logi TRUE
.. .. .. .. .. .. .. ..$ latent_var : chr "log_umi"
.. .. .. .. .. .. .. ..$ batch_var : NULL
.. .. .. .. .. .. .. ..$ latent_var_nonreg : NULL
.. .. .. .. .. .. .. ..$ n_genes : num 2000
.. .. .. .. .. .. .. ..$ n_cells : num 491
.. .. .. .. .. .. .. ..$ method : chr "glmGamPoi_offset"
.. .. .. .. .. .. .. ..$ do_regularize : logi TRUE
.. .. .. .. .. .. .. ..$ theta_regularization : chr "od_factor"
.. .. .. .. .. .. .. ..$ res_clip_range : num [1:2] -22.2 22.2
.. .. .. .. .. .. .. ..$ bin_size : num 500
.. .. .. .. .. .. .. ..$ min_cells : num 5
.. .. .. .. .. .. .. ..$ residual_type : chr "pearson"
.. .. .. .. .. .. .. ..$ return_cell_attr : logi TRUE
.. .. .. .. .. .. .. ..$ return_gene_attr : logi TRUE
.. .. .. .. .. .. .. ..$ return_corrected_umi : logi TRUE
.. .. .. .. .. .. .. ..$ min_variance : chr "umi_median"
.. .. .. .. .. .. .. ..$ bw_adjust : num 3
.. .. .. .. .. .. .. ..$ gmean_eps : num 1
.. .. .. .. .. .. .. ..$ theta_estimation_fun : chr "theta.ml"
.. .. .. .. .. .. .. ..$ theta_given : NULL
.. .. .. .. .. .. .. ..$ exclude_poisson : logi TRUE
.. .. .. .. .. .. .. ..$ use_geometric_mean : logi TRUE
.. .. .. .. .. .. .. ..$ use_geometric_mean_offset: logi FALSE
.. .. .. .. .. .. .. ..$ fix_intercept : logi FALSE
.. .. .. .. .. .. .. ..$ fix_slope : logi FALSE
.. .. .. .. .. .. .. ..$ scale_factor : logi NA
.. .. .. .. .. .. .. ..$ vst.flavor : chr "v2"
.. .. .. .. .. .. .. ..$ verbosity : num 0
.. .. .. .. .. .. .. ..$ verbose : NULL
.. .. .. .. .. .. .. ..$ show_progress : NULL
.. .. .. .. .. .. .. ..$ set_min_var : num 0.16
.. .. .. .. .. .. .. ..$ sct.clip.range : num [1:2] -4.05 4.05
.. .. .. .. .. .. ..@ median_umi : num 4153
.. .. .. ..@ counts :Formal class 'dgCMatrix' [package "Matrix"] with 6 slots
.. .. .. .. .. ..@ i : int [1:429533] 14 61 73 107 147 187 192 223 244 250 ...
.. .. .. .. .. ..@ p : int [1:492] 0 644 1219 2551 3098 3508 4745 6165 7529 8150 ...
.. .. .. .. .. ..@ Dim : int [1:2] 12084 491
.. .. .. .. .. ..@ Dimnames:List of 2
.. .. .. .. .. .. ..$ : chr [1:12084] "Gene0000010" "Gene0000020" "Gene0000030" "Gene0000040" ...
.. .. .. .. .. .. ..$ : chr [1:491] "36077725294100" "36077725294200" "36077725294300" "36077725294400" ...
.. .. .. .. .. ..@ x : num [1:429533] 10 3 2 5 1 1 1 1 4 1 ...
.. .. .. .. .. ..@ factors : list()
.. .. .. ..@ data :Formal class 'dgCMatrix' [package "Matrix"] with 6 slots
.. .. .. .. .. ..@ i : int [1:429533] 14 61 73 107 147 187 192 223 244 250 ...
.. .. .. .. .. ..@ p : int [1:492] 0 644 1219 2551 3098 3508 4745 6165 7529 8150 ...
.. .. .. .. .. ..@ Dim : int [1:2] 12084 491
.. .. .. .. .. ..@ Dimnames:List of 2
.. .. .. .. .. .. ..$ : chr [1:12084] "Gene0000010" "Gene0000020" "Gene0000030" "Gene0000040" ...
.. .. .. .. .. .. ..$ : chr [1:491] "36077725294100" "36077725294200" "36077725294300" "36077725294400" ...
.. .. .. .. .. ..@ x : num [1:429533] 3.54 2.4 2.04 2.87 1.47 ...
.. .. .. .. .. ..@ factors : list()
.. .. .. ..@ scale.data : num [1:2000, 1:491] -0.212 -0.504 -0.386 -0.251 0.41 ...
.. .. .. .. ..- attr(*, "dimnames")=List of 2
.. .. .. .. .. ..$ : chr [1:2000] "Gene0000010" "Gene0000040" "Gene0000080" "Gene0000130" ...
.. .. .. .. .. ..$ : chr [1:491] "36077725294100" "36077725294200" "36077725294300" "36077725294400" ...
.. .. .. ..@ assay.orig : chr "Spatial"
.. .. .. ..@ var.features : chr [1:2000] "Gene0062990" "Gene0061940" "Gene0118390" "Gene0327200" ...
.. .. .. ..@ meta.features:'data.frame': 12084 obs. of 0 variables
.. .. .. ..@ misc : Named list()
.. .. .. ..@ key : chr "sct_"
..@ meta.data :'data.frame': 491 obs. of 18 variables:
.. ..$ Row.names : 'AsIs' chr [1:491] "36077725294100" "36077725294200" "36077725294300" "36077725294400" ...
.. ..$ _index : chr [1:491] "36077725294100" "36077725294200" "36077725294300" "36077725294400" ...
.. ..$ nCount_Spatial : num [1:491] 32 2966 7874 2277 1121 ...
.. ..$ nFeature_Spatial: int [1:491] 21 584 1441 555 330 1278 2005 1497 63 391 ...
.. ..$ orig.ident : chr [1:491] "A" "A" "A" "A" ...
.. ..$ percent.mito : num [1:491] 0 0 0 0 0 0 0 0 0 0 ...
.. ..$ x : int [1:491] 8400 8400 8400 8400 8500 8500 8500 8500 8600 8600 ...
.. ..$ y : int [1:491] 7700 7800 7900 8000 7700 7800 7900 8000 6700 6800 ...
.. ..$ nCount_SCT : num [1:491] 2995 3923 4438 3790 3493 ...
.. ..$ nFeature_SCT : int [1:491] 644 575 1332 547 410 1237 1420 1364 621 446 ...
.. ..$ SCT_snn_res.1.2 : Factor w/ 7 levels "0","1","2","3",..: 2 5 1 1 2 1 1 1 2 2 ...
.. ..$ seurat_clusters : Factor w/ 5 levels "0","1","2","3",..: 5 1 1 1 3 1 1 1 5 1 ...
.. ..$ tissue : num [1:491] 1 1 1 1 1 1 1 1 1 1 ...
.. ..$ row : num [1:491] 5201 5301 5401 5501 5201 ...
.. ..$ col : num [1:491] 3201 3201 3201 3201 3301 ...
.. ..$ imagerow : num [1:491] 5201 5301 5401 5501 5201 ...
.. ..$ imagecol : num [1:491] 3201 3201 3201 3201 3301 ...
.. ..$ SCT_snn_res.0.8 : Factor w/ 5 levels "0","1","2","3",..: 5 1 1 1 3 1 1 1 5 1 ...
..@ active.assay: chr "SCT"
..@ active.ident: Factor w/ 5 levels "0","1","2","3",..: 5 1 1 1 3 1 1 1 5 1 ...
.. ..- attr(*, "names")= chr [1:491] "36077725294100" "36077725294200" "36077725294300" "36077725294400" ...
..@ graphs :List of 2
.. ..$ SCT_nn :Formal class 'Graph' [package "SeuratObject"] with 7 slots
.. .. .. ..@ assay.used: chr "SCT"
.. .. .. ..@ i : int [1:9820] 0 8 37 54 74 95 234 235 236 237 ...
.. .. .. ..@ p : int [1:492] 0 21 55 62 74 138 159 168 183 210 ...
.. .. .. ..@ Dim : int [1:2] 491 491
.. .. .. ..@ Dimnames :List of 2
.. .. .. .. ..$ : chr [1:491] "36077725294100" "36077725294200" "36077725294300" "36077725294400" ...
.. .. .. .. ..$ : chr [1:491] "36077725294100" "36077725294200" "36077725294300" "36077725294400" ...
.. .. .. ..@ x : num [1:9820] 1 1 1 1 1 1 1 1 1 1 ...
.. .. .. ..@ factors : list()
.. ..$ SCT_snn:Formal class 'Graph' [package "SeuratObject"] with 7 slots
.. .. .. ..@ assay.used: chr "SCT"
.. .. .. ..@ i : int [1:39087] 0 8 12 15 16 37 54 74 95 117 ...
.. .. .. ..@ p : int [1:492] 0 32 143 229 344 431 534 586 647 679 ...
.. .. .. ..@ Dim : int [1:2] 491 491
.. .. .. ..@ Dimnames :List of 2
.. .. .. .. ..$ : chr [1:491] "36077725294100" "36077725294200" "36077725294300" "36077725294400" ...
.. .. .. .. ..$ : chr [1:491] "36077725294100" "36077725294200" "36077725294300" "36077725294400" ...
.. .. .. ..@ x : num [1:39087] 1 1 0.29 0.379 0.333 ...
.. .. .. ..@ factors : list()
..@ neighbors : list()
..@ reductions :List of 2
.. ..$ pca :Formal class 'DimReduc' [package "SeuratObject"] with 9 slots
.. .. .. ..@ cell.embeddings : num [1:491, 1:50] -0.321 7.57 11.047 8.912 5.363 ...
.. .. .. .. ..- attr(*, "dimnames")=List of 2
.. .. .. .. .. ..$ : chr [1:491] "36077725294100" "36077725294200" "36077725294300" "36077725294400" ...
.. .. .. .. .. ..$ : chr [1:50] "PC_1" "PC_2" "PC_3" "PC_4" ...
.. .. .. ..@ feature.loadings : num [1:2000, 1:50] -0.0706 -0.0691 -0.0804 -0.077 -0.0722 ...
.. .. .. .. ..- attr(*, "dimnames")=List of 2
.. .. .. .. .. ..$ : chr [1:2000] "Gene0062990" "Gene0061940" "Gene0118390" "Gene0327200" ...
.. .. .. .. .. ..$ : chr [1:50] "PC_1" "PC_2" "PC_3" "PC_4" ...
.. .. .. ..@ feature.loadings.projected: num[0 , 0 ]
.. .. .. ..@ assay.used : chr "SCT"
.. .. .. ..@ global : logi FALSE
.. .. .. ..@ stdev : num [1:50] 8.38 7.7 4.84 4.04 3.89 ...
.. .. .. ..@ jackstraw :Formal class 'JackStrawData' [package "SeuratObject"] with 4 slots
.. .. .. .. .. ..@ empirical.p.values : num[0 , 0 ]
.. .. .. .. .. ..@ fake.reduction.scores : num[0 , 0 ]
.. .. .. .. .. ..@ empirical.p.values.full: num[0 , 0 ]
.. .. .. .. .. ..@ overall.p.values : num[0 , 0 ]
.. .. .. ..@ misc :List of 1
.. .. .. .. ..$ total.variance: num 1994
.. .. .. ..@ key : chr "PC_"
.. ..$ umap:Formal class 'DimReduc' [package "SeuratObject"] with 9 slots
.. .. .. ..@ cell.embeddings : num [1:491, 1:2] -4.518 -2.5526 -0.0788 -1.302 -3.5134 ...
.. .. .. .. ..- attr(*, "scaled:center")= num [1:2] -7.54 -5.65
.. .. .. .. ..- attr(*, "dimnames")=List of 2
.. .. .. .. .. ..$ : chr [1:491] "36077725294100" "36077725294200" "36077725294300" "36077725294400" ...
.. .. .. .. .. ..$ : chr [1:2] "umap_1" "umap_2"
.. .. .. ..@ feature.loadings : num[0 , 0 ]
.. .. .. ..@ feature.loadings.projected: num[0 , 0 ]
.. .. .. ..@ assay.used : chr "SCT"
.. .. .. ..@ global : logi TRUE
.. .. .. ..@ stdev : num(0)
.. .. .. ..@ jackstraw :Formal class 'JackStrawData' [package "SeuratObject"] with 4 slots
.. .. .. .. .. ..@ empirical.p.values : num[0 , 0 ]
.. .. .. .. .. ..@ fake.reduction.scores : num[0 , 0 ]
.. .. .. .. .. ..@ empirical.p.values.full: num[0 , 0 ]
.. .. .. .. .. ..@ overall.p.values : num[0 , 0 ]
.. .. .. ..@ misc : list()
.. .. .. ..@ key : chr "umap_"
..@ images :List of 1
.. ..$ slice1:Formal class 'VisiumV1' [package "Seurat"] with 6 slots
.. .. .. ..@ image : num [1:6101, 1:5901] 1 1 1 1 1 1 1 1 1 1 ...
.. .. .. ..@ scale.factors:List of 4
.. .. .. .. ..$ spot : num 1
.. .. .. .. ..$ fiducial: num 1
.. .. .. .. ..$ hires : num 1
.. .. .. .. ..$ lowres : num 1
.. .. .. .. ..- attr(*, "class")= chr "scalefactors"
.. .. .. ..@ coordinates :'data.frame': 491 obs. of 5 variables:
.. .. .. .. ..$ tissue : num [1:491] 1 1 1 1 1 1 1 1 1 1 ...
.. .. .. .. ..$ row : num [1:491] 5201 5301 5401 5501 5201 ...
.. .. .. .. ..$ col : num [1:491] 3201 3201 3201 3201 3301 ...
.. .. .. .. ..$ imagerow: num [1:491] 5201 5301 5401 5501 5201 ...
.. .. .. .. ..$ imagecol: num [1:491] 3201 3201 3201 3201 3301 ...
.. .. .. ..@ spot.radius : num 0.000164
.. .. .. ..@ assay : chr "Spatial"
.. .. .. ..@ key : chr "slice1_"
..@ project.name: chr "AnnData"
..@ misc :List of 6
.. ..$ raw_cellname: chr [1:984] "22333829942000" "22333829942100" "22333829942200" "22333829942300" ...
.. ..$ raw_counts :Formal class 'dgCMatrix' [package "Matrix"] with 6 slots
.. .. .. ..@ i : int [1:965369] 14 77 92 142 218 272 350 405 492 560 ...
.. .. .. ..@ p : int [1:985] 0 253 936 1901 2909 3419 3465 3553 4323 5593 ...
.. .. .. ..@ Dim : int [1:2] 20092 984
.. .. .. ..@ Dimnames:List of 2
.. .. .. .. ..$ : NULL
.. .. .. .. ..$ : NULL
.. .. .. ..@ x : num [1:965369] 3 1 1 1 1 1 2 1 3 1 ...
.. .. .. ..@ factors : list()
.. ..$ raw_genename: chr [1:20092] "Gene0000010" "Gene0000020" "Gene0000030" "Gene0000040" ...
.. ..$ sn :List of 3
.. .. ..$ _index: int 0
.. .. ..$ batch : chr "-1"
.. .. ..$ sn : chr "B03203E412"
.. ..$ active_wgcna: chr "vis"
.. ..$ vis :List of 1
.. .. ..$ wgcna_genes: chr [1:4058] "Gene0000040" "Gene0000050" "Gene0000070" "Gene0000080" ...
..@ version :Classes 'package_version', 'numeric_version' hidden list of 1
.. ..$ : int [1:4] 3 1 5 9900
..@ commands :List of 9
.. ..$ NormalizeData.Spatial :Formal class 'SeuratCommand' [package "SeuratObject"] with 5 slots
.. .. .. ..@ name : chr "NormalizeData.Spatial"
.. .. .. ..@ time.stamp : POSIXct[1:1], format: "2024-03-18 12:11:13"
.. .. .. ..@ assay.used : chr "Spatial"
.. .. .. ..@ call.string: chr "NormalizeData(object)"
.. .. .. ..@ params :List of 5
.. .. .. .. ..$ assay : chr "Spatial"
.. .. .. .. ..$ normalization.method: chr "LogNormalize"
.. .. .. .. ..$ scale.factor : num 10000
.. .. .. .. ..$ margin : num 1
.. .. .. .. ..$ verbose : logi TRUE
.. ..$ SCTransform.Spatial :Formal class 'SeuratCommand' [package "SeuratObject"] with 5 slots
.. .. .. ..@ name : chr "SCTransform.Spatial"
.. .. .. ..@ time.stamp : POSIXct[1:1], format: "2024-04-08 19:49:28"
.. .. .. ..@ assay.used : chr "Spatial"
.. .. .. ..@ call.string: chr "SCTransform(tA, assay = \"Spatial\", verbose = FALSE)"
.. .. .. ..@ params :List of 14
.. .. .. .. ..$ assay : chr "Spatial"
.. .. .. .. ..$ new.assay.name : chr "SCT"
.. .. .. .. ..$ do.correct.umi : logi TRUE
.. .. .. .. ..$ ncells : num 5000
.. .. .. .. ..$ variable.features.n : num 3000
.. .. .. .. ..$ variable.features.rv.th: num 1.3
.. .. .. .. ..$ do.scale : logi FALSE
.. .. .. .. ..$ do.center : logi TRUE
.. .. .. .. ..$ clip.range : num [1:2] -4.05 4.05
.. .. .. .. ..$ vst.flavor : chr "v2"
.. .. .. .. ..$ conserve.memory : logi FALSE
.. .. .. .. ..$ return.only.var.genes : logi TRUE
.. .. .. .. ..$ seed.use : num 1448145
.. .. .. .. ..$ verbose : logi FALSE
.. ..$ NormalizeData.SCT :Formal class 'SeuratCommand' [package "SeuratObject"] with 5 slots
.. .. .. ..@ name : chr "NormalizeData.SCT"
.. .. .. ..@ time.stamp : POSIXct[1:1], format: "2024-04-08 19:57:51"
.. .. .. ..@ assay.used : chr "SCT"
.. .. .. ..@ call.string: chr "NormalizeData(.)"
.. .. .. ..@ params :List of 5
.. .. .. .. ..$ assay : chr "SCT"
.. .. .. .. ..$ normalization.method: chr "LogNormalize"
.. .. .. .. ..$ scale.factor : num 10000
.. .. .. .. ..$ margin : num 1
.. .. .. .. ..$ verbose : logi TRUE
.. ..$ FindVariableFeatures.SCT:Formal class 'SeuratCommand' [package "SeuratObject"] with 5 slots
.. .. .. ..@ name : chr "FindVariableFeatures.SCT"
.. .. .. ..@ time.stamp : POSIXct[1:1], format: "2024-04-08 19:57:51"
.. .. .. ..@ assay.used : chr "SCT"
.. .. .. ..@ call.string: chr "FindVariableFeatures(.)"
.. .. .. ..@ params :List of 12
.. .. .. .. ..$ assay : chr "SCT"
.. .. .. .. ..$ selection.method : chr "vst"
.. .. .. .. ..$ loess.span : num 0.3
.. .. .. .. ..$ clip.max : chr "auto"
.. .. .. .. ..$ mean.function :function (mat, display_progress)
.. .. .. .. ..$ dispersion.function:function (mat, display_progress)
.. .. .. .. ..$ num.bin : num 20
.. .. .. .. ..$ binning.method : chr "equal_width"
.. .. .. .. ..$ nfeatures : num 2000
.. .. .. .. ..$ mean.cutoff : num [1:2] 0.1 8
.. .. .. .. ..$ dispersion.cutoff : num [1:2] 1 Inf
.. .. .. .. ..$ verbose : logi TRUE
.. ..$ ScaleData.SCT :Formal class 'SeuratCommand' [package "SeuratObject"] with 5 slots
.. .. .. ..@ name : chr "ScaleData.SCT"
.. .. .. ..@ time.stamp : POSIXct[1:1], format: "2024-04-08 19:57:51"
.. .. .. ..@ assay.used : chr "SCT"
.. .. .. ..@ call.string: chr "ScaleData(.)"
.. .. .. ..@ params :List of 10
.. .. .. .. ..$ features : chr [1:2000] "Gene0062990" "Gene0061940" "Gene0118390" "Gene0327200" ...
.. .. .. .. ..$ assay : chr "SCT"
.. .. .. .. ..$ model.use : chr "linear"
.. .. .. .. ..$ use.umi : logi FALSE
.. .. .. .. ..$ do.scale : logi TRUE
.. .. .. .. ..$ do.center : logi TRUE
.. .. .. .. ..$ scale.max : num 10
.. .. .. .. ..$ block.size : num 1000
.. .. .. .. ..$ min.cells.to.block: num 491
.. .. .. .. ..$ verbose : logi TRUE
.. ..$ RunPCA.SCT :Formal class 'SeuratCommand' [package "SeuratObject"] with 5 slots
.. .. .. ..@ name : chr "RunPCA.SCT"
.. .. .. ..@ time.stamp : POSIXct[1:1], format: "2024-04-08 19:57:52"
.. .. .. ..@ assay.used : chr "SCT"
.. .. .. ..@ call.string: chr "RunPCA(.)"
.. .. .. ..@ params :List of 10
.. .. .. .. ..$ assay : chr "SCT"
.. .. .. .. ..$ npcs : num 50
.. .. .. .. ..$ rev.pca : logi FALSE
.. .. .. .. ..$ weight.by.var : logi TRUE
.. .. .. .. ..$ verbose : logi TRUE
.. .. .. .. ..$ ndims.print : int [1:5] 1 2 3 4 5
.. .. .. .. ..$ nfeatures.print: num 30
.. .. .. .. ..$ reduction.name : chr "pca"
.. .. .. .. ..$ reduction.key : chr "PC_"
.. .. .. .. ..$ seed.use : num 42
.. ..$ FindNeighbors.SCT.pca :Formal class 'SeuratCommand' [package "SeuratObject"] with 5 slots
.. .. .. ..@ name : chr "FindNeighbors.SCT.pca"
.. .. .. ..@ time.stamp : POSIXct[1:1], format: "2024-04-08 19:57:53"
.. .. .. ..@ assay.used : chr "SCT"
.. .. .. ..@ call.string: chr "FindNeighbors(ta_obj, dims = 1:30)"
.. .. .. ..@ params :List of 16
.. .. .. .. ..$ reduction : chr "pca"
.. .. .. .. ..$ dims : int [1:30] 1 2 3 4 5 6 7 8 9 10 ...
.. .. .. .. ..$ assay : chr "SCT"
.. .. .. .. ..$ k.param : num 20
.. .. .. .. ..$ return.neighbor: logi FALSE
.. .. .. .. ..$ compute.SNN : logi TRUE
.. .. .. .. ..$ prune.SNN : num 0.0667
.. .. .. .. ..$ nn.method : chr "annoy"
.. .. .. .. ..$ n.trees : num 50
.. .. .. .. ..$ annoy.metric : chr "euclidean"
.. .. .. .. ..$ nn.eps : num 0
.. .. .. .. ..$ verbose : logi TRUE
.. .. .. .. ..$ do.plot : logi FALSE
.. .. .. .. ..$ graph.name : chr [1:2] "SCT_nn" "SCT_snn"
.. .. .. .. ..$ l2.norm : logi FALSE
.. .. .. .. ..$ cache.index : logi FALSE
.. ..$ FindClusters :Formal class 'SeuratCommand' [package "SeuratObject"] with 5 slots
.. .. .. ..@ name : chr "FindClusters"
.. .. .. ..@ time.stamp : POSIXct[1:1], format: "2024-04-08 19:57:53"
.. .. .. ..@ assay.used : chr "SCT"
.. .. .. ..@ call.string: chr "FindClusters(ta_obj, verbose = TRUE)"
.. .. .. ..@ params :List of 11
.. .. .. .. ..$ graph.name : chr "SCT_snn"
.. .. .. .. ..$ cluster.name : chr "SCT_snn_res.0.8"
.. .. .. .. ..$ modularity.fxn : num 1
.. .. .. .. ..$ resolution : num 0.8
.. .. .. .. ..$ method : chr "matrix"
.. .. .. .. ..$ algorithm : num 1
.. .. .. .. ..$ n.start : num 10
.. .. .. .. ..$ n.iter : num 10
.. .. .. .. ..$ random.seed : num 0
.. .. .. .. ..$ group.singletons: logi TRUE
.. .. .. .. ..$ verbose : logi TRUE
.. ..$ RunUMAP.SCT.pca :Formal class 'SeuratCommand' [package "SeuratObject"] with 5 slots
.. .. .. ..@ name : chr "RunUMAP.SCT.pca"
.. .. .. ..@ time.stamp : POSIXct[1:1], format: "2024-04-08 19:57:57"
.. .. .. ..@ assay.used : chr "SCT"
.. .. .. ..@ call.string: chr "RunUMAP(ta_obj, dims = 1:30)"
.. .. .. ..@ params :List of 25
.. .. .. .. ..$ dims : int [1:30] 1 2 3 4 5 6 7 8 9 10 ...
.. .. .. .. ..$ reduction : chr "pca"
.. .. .. .. ..$ assay : chr "SCT"
.. .. .. .. ..$ slot : chr "data"
.. .. .. .. ..$ umap.method : chr "uwot"
.. .. .. .. ..$ return.model : logi FALSE
.. .. .. .. ..$ n.neighbors : int 30
.. .. .. .. ..$ n.components : int 2
.. .. .. .. ..$ metric : chr "cosine"
.. .. .. .. ..$ learning.rate : num 1
.. .. .. .. ..$ min.dist : num 0.3
.. .. .. .. ..$ spread : num 1
.. .. .. .. ..$ set.op.mix.ratio : num 1
.. .. .. .. ..$ local.connectivity : int 1
.. .. .. .. ..$ repulsion.strength : num 1
.. .. .. .. ..$ negative.sample.rate: int 5
.. .. .. .. ..$ uwot.sgd : logi FALSE
.. .. .. .. ..$ seed.use : int 42
.. .. .. .. ..$ angular.rp.forest : logi FALSE
.. .. .. .. ..$ densmap : logi FALSE
.. .. .. .. ..$ dens.lambda : num 2
.. .. .. .. ..$ dens.frac : num 0.3
.. .. .. .. ..$ dens.var.shift : num 0.1
.. .. .. .. ..$ verbose : logi TRUE
.. .. .. .. ..$ reduction.name : chr "umap"
..@ tools : Named list()
Describe the bug When I use MetaspotsByGroups to treat my seurat_obj, It reports error. Looks like follow:
After I tired to debug. I found the problem may be from followed code ( https://github.com/smorabit/hdWGCNA/blob/b3b5acf3f1cff5fd702513ebb7e43545c6bcd725/R/Metaspots.R#L80) :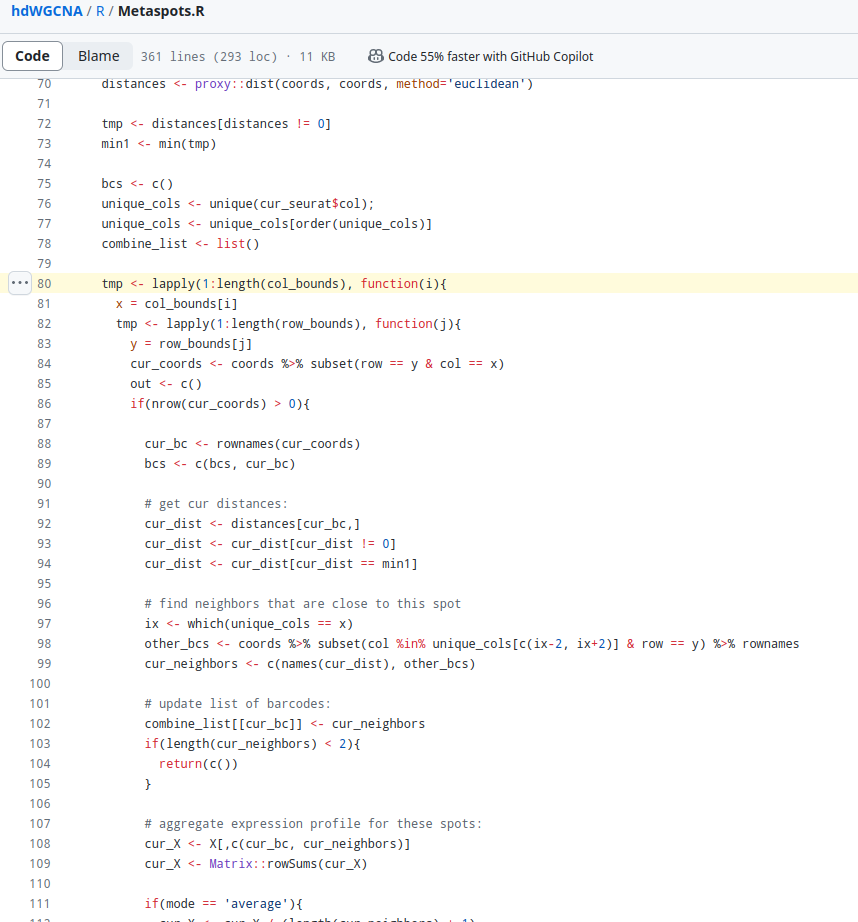
In my test, I always get empty element with
cur_coordsSadly, It's so hard for me to deeply debug , I hope I could run this app.R session info