Hi,
This is a very interesting question, but at this time I believe that it is outside of the scope of hdWGCNA since you are essentially wanting to calculate a custom metric. I do not know anything about this particular calculation so I don't think I can provide meaningful advice. It seems like you can just write a custom function to implement this calculation for your purposes.
Hi Sam, I'm currently making metacells within each condition and identifying modules over all conditions, then doing DME. I want to compare the connectivity of module in each condition, defined as the ratio of sum of all edge weight if I understood correctly this passage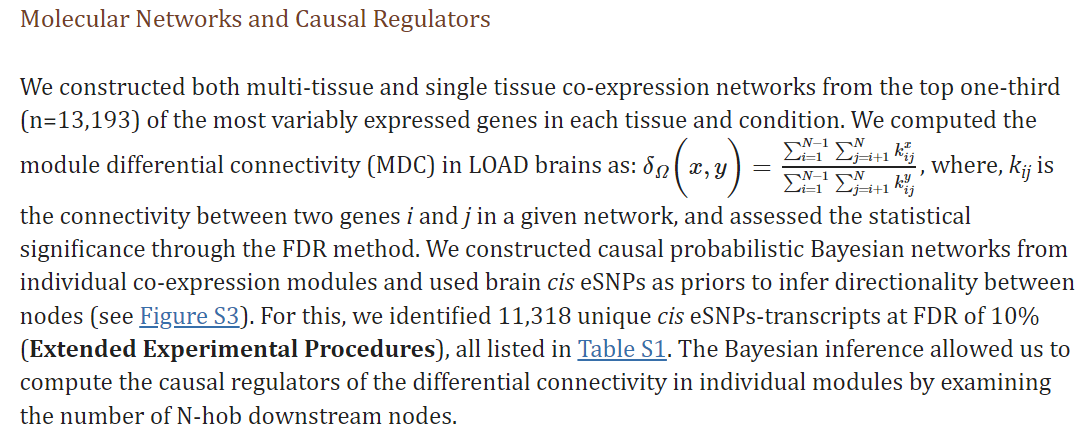 from this paper:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3677161/
from this paper:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3677161/
Since I constructed the network over all conditions I assume the edge weight in TOM is also over all conditions. Would it be a good idea to do the following: a) subset the finished WGCNA/Seurat object by condition to keep the metacell composition b) construct the networks c) extract edge weight using the previously defined network from TOM
I also wonder if I should keep all edges in the sum, or only those are positive?
Any suggestions are welcome! Thank you!