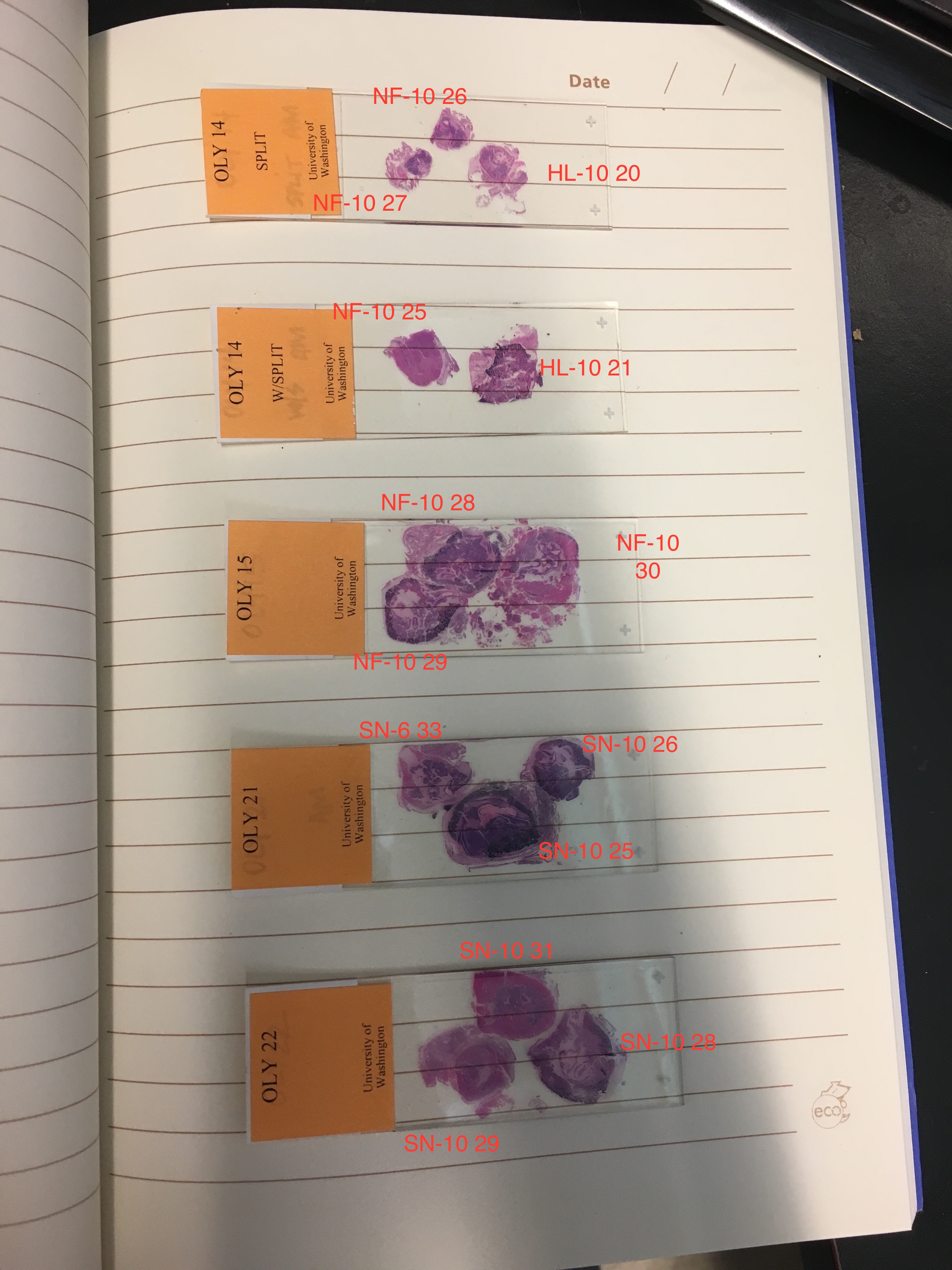
Final list:
SN low pH
- male - SN-10-20
- female - SN-10-16
- hermaphrodite - SN-10-17
SN ambient pH
- male - SN-10-25
- female - SN-10-31
- hermaphrodite - SN-10-26
NF low pH
- male - NF-10-24
- female - NF-10-23
- hermaphrodite - NF-10 22
NF ambient pH
- male - NF-10-28 (not 100% male, a few oocytes)
- female - NF-10-30
- hermaphrodite - NF-10-26
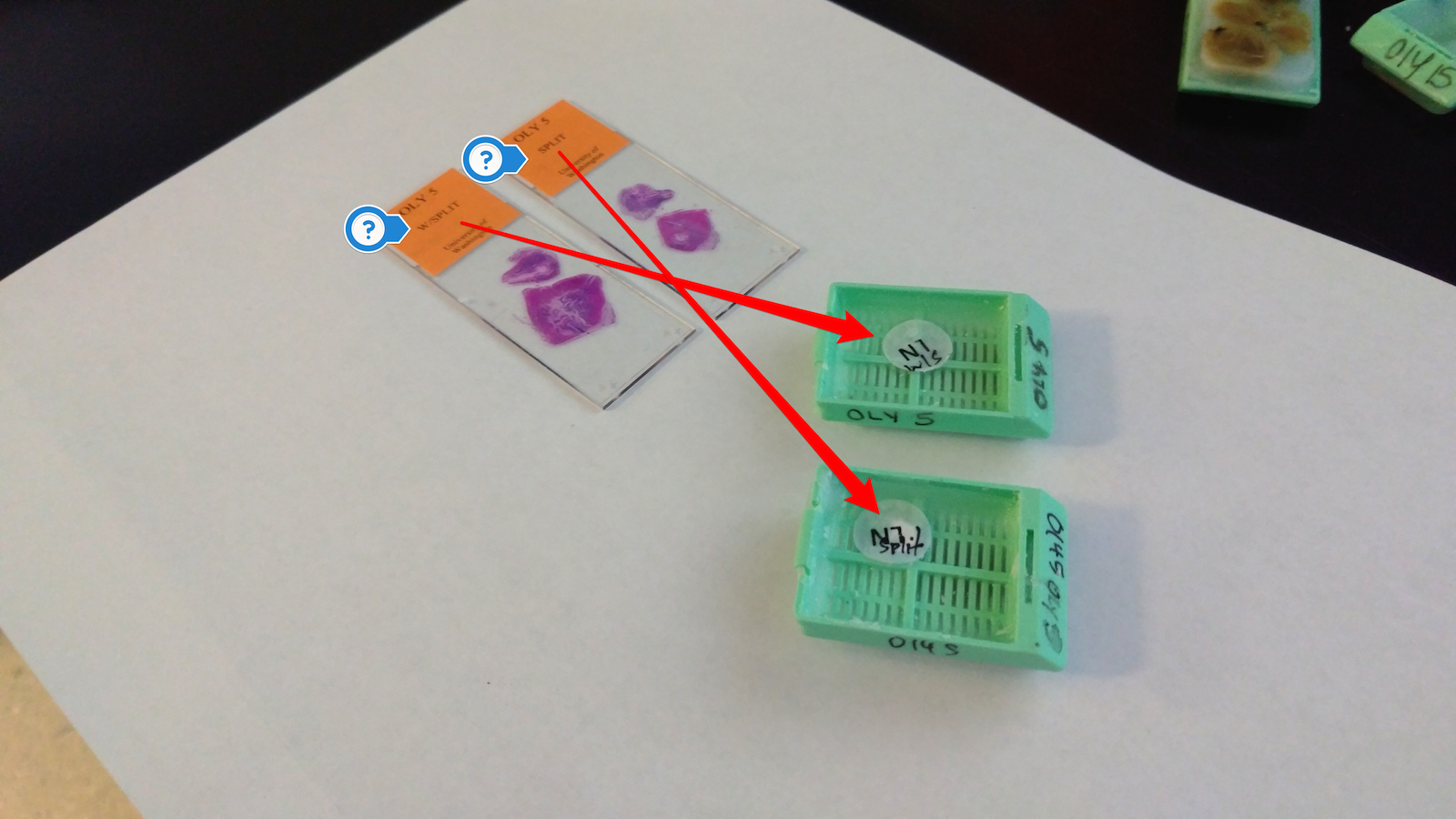
@kubu4, you'll probably want to work with Katie to make sure that you're extracting from the same sample as the one she imaged. The histology folks did not keep the samples in the same formation. Katie used my images of the prepared cassettes and my histology "map" to ID each sample.








For Katherine's libraries..
(Paxgene kit is here)
Samples are by scope in lab.
SN low pH male - SN-10-20 female - SN-10-16 hermaphrodite - SN-10-17
SN ambient pH
NF low pH
NF ambient pH - (missing image for NF-10-25)