Just to reiterate and make it clear for anyone else - using the standalone R software is preferred .
I think this is some issue with rpy2.
I suggest trying in R first, make sure it works, then if you want, you could troubleshoot (but not necessary to get it to run in Jupyter.
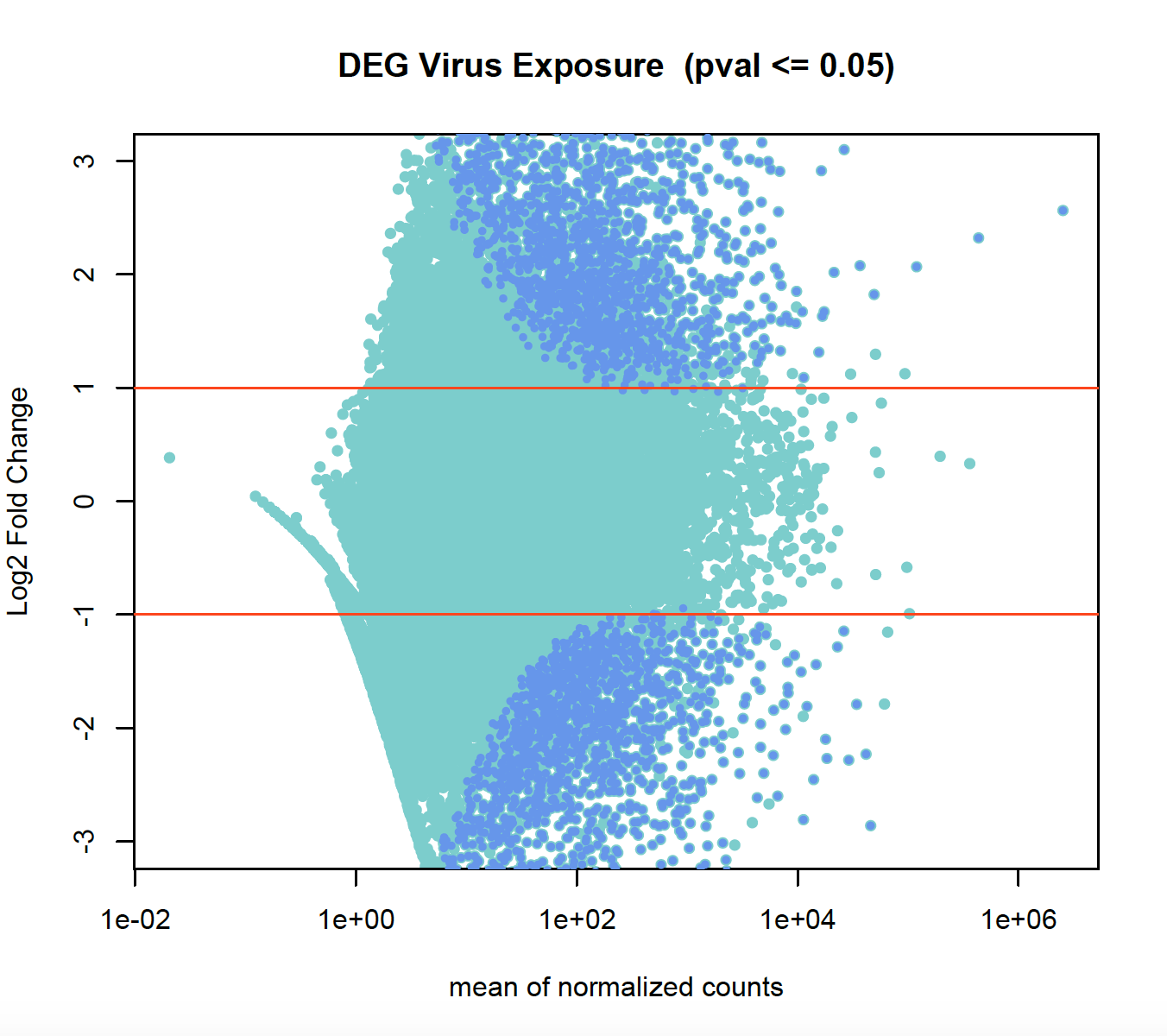
I don't use R, so there is probably an obvious problem with what I'm trying to do here... I'm trying to run the R script for the quiz in an R Jupyter notebook.
What I've done so far: I installed R and got rpy2 in this notebook. This seemed to go fine. I downloaded the
Phel_countdata.txtfile in another notebook.Then I started an R notebook to run the script needed, as you can see here. But I can't get to DESeq2 to install correctly, hence this error message:
If case you don't want to follow the R nb link, a screenshot: