fuzznuc is a tool that finds motifs (in this case your are looking for all CG motifs in your sequence.
I believe Galaxy has this tool, you can run it from the command-line, and there are various other web sites that host it.
ie http://www.bioinformatics.nl/cgi-bin/emboss/fuzznuc
There are several output types, but you might want GFF for browser..
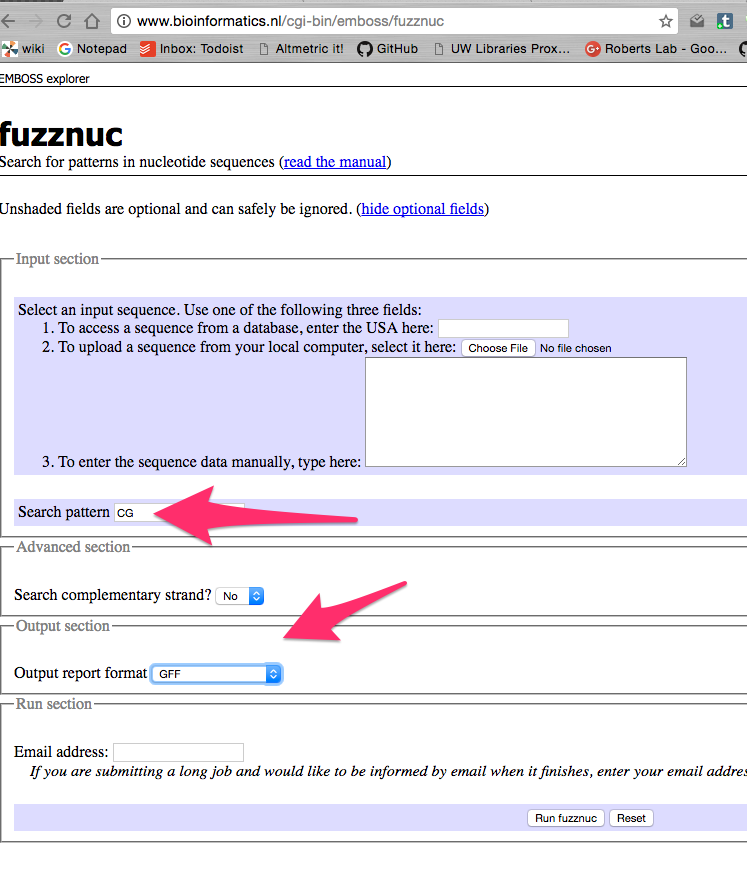
I'm seeking to identify potential methylation sites (CpG's) in my geoduck genome; although the CpG sites aren't definitive methylation sites, they have potential to be. I believe CoGe only has programs to identify methylation in bisulfite treated RNASeq files (FASTQ), and it doesn't allow one to select a FASTA file. CoGe format options are: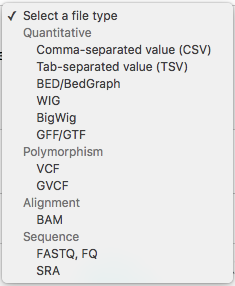
Does anyone have suggestions on a program/script to use to identify CpG sites, or possibly "islands", in a genome? Thanks!