What’s the architecture being considered here? It looks like you’re having each sampler fill the parallel chains individually which is a huge duplication of code and not a robust way to approach is statistically.
On Jul 7, 2014, at 7:43 PM, Peter Li notifications@github.com wrote:
Need to change sample to std::vector in ....
src/stan/agrad/rev/matrix/sd.hpp src/stan/agrad/rev/matrix/variance.hpp src/stan/command/print.cpp src/stan/common/command.hpp src/stan/common/init_adapt.hpp src/stan/common/init_nuts.hpp src/stan/common/init_static_hmc.hpp src/stan/common/init_windowed_adapt.hpp src/stan/common/run_markov_chain.hpp src/stan/common/sample.hpp src/stan/common/warmup.hpp src/stan/common/write_error_msg.hpp src/stan/common.hpp src/stan/gm/arguments/arg_method.hpp src/stan/gm/arguments/arg_num_samples.hpp src/stan/gm/arguments/arg_sample.hpp src/stan/gm/arguments/arg_sample_algo.hpp src/stan/gm/arguments/arg_save_warmup.hpp src/stan/gm/arguments/arg_thin.hpp src/stan/gm/ast.hpp src/stan/gm/ast_def.cpp src/stan/gm/generator.hpp src/stan/gm/grammars/statement_2_grammar_def.hpp src/stan/gm/grammars/statement_grammar.hpp src/stan/gm/grammars/statement_grammar_def.hpp src/stan/io/mcmc_writer.hpp src/stan/io/stan_csv_reader.hpp src/stan/math/functions/Phi.hpp src/stan/math/functions/Phi_approx.hpp src/stan/math/matrix/mean.hpp src/stan/math/matrix/sd.hpp src/stan/math/matrix/variance.hpp src/stan/mcmc/base_mcmc.hpp src/stan/mcmc/chains.hpp src/stan/mcmc/covar_adaptation.hpp src/stan/mcmc/fixed_param_sampler.hpp src/stan/mcmc/hmc/base_hmc.hpp src/stan/mcmc/hmc/hamiltonians/base_hamiltonian.hpp src/stan/mcmc/hmc/hamiltonians/dense_e_metric.hpp src/stan/mcmc/hmc/hamiltonians/diag_e_metric.hpp src/stan/mcmc/hmc/hamiltonians/unit_e_metric.hpp src/stan/mcmc/hmc/nuts/adapt_dense_e_nuts.hpp src/stan/mcmc/hmc/nuts/adapt_diag_e_nuts.hpp src/stan/mcmc/hmc/nuts/adapt_unit_e_nuts.hpp src/stan/mcmc/hmc/nuts/base_nuts.hpp src/stan/mcmc/hmc/static/adapt_dense_e_static_hmc.hpp src/stan/mcmc/hmc/static/adapt_diag_e_static_hmc.hpp src/stan/mcmc/hmc/static/adapt_unit_e_static_hmc.hpp src/stan/mcmc/hmc/static/base_static_hmc.hpp src/stan/mcmc/sample.hpp src/stan/mcmc/var_adaptation.hpp src/stan/mcmc.hpp src/stan/prob/distributions/multivariate/continuous/dirichlet.hpp src/stan/prob/distributions/univariate/continuous/beta.hpp src/stan/prob/distributions/univariate/discrete/beta_binomial.hpp src/stan/prob/welford_covar_estimator.hpp src/stan/prob/welford_var_estimator.hpp
src/test/test-models/syntax-only/function_signatures1.stan
src/test/unit/common/command_init_test.cpp src/test/unit/common/do_bfgs_optimize_test.cpp src/test/unit/common/run_markov_chain_test.cpp src/test/unit/common/sample_test.cpp src/test/unit/common/warmup_test.cpp src/test/unit/distribution/multivariate/continuous/dirichlet_test.cpp src/test/unit/gm/parser_test.cpp src/test/unit/io/mcmc_writer_test.cpp src/test/unit/io/stan_csv_reader_test.cpp src/test/unit/io/test_csv_files/blocker.0.csv src/test/unit/io/test_csv_files/blocker_nondiag.0.csv src/test/unit/io/test_csv_files/epil.0.csv src/test/unit/io/test_csv_files/metadata1.csv src/test/unit/io/test_csv_files/metadata2.csv src/test/unit/mcmc/chains_test.cpp src/test/unit/mcmc/hmc/base_hmc_test.cpp src/test/unit/mcmc/hmc/base_nuts_test.cpp src/test/unit/mcmc/hmc/base_static_hmc_test.cpp src/test/unit/mcmc/hmc/derived_nuts_test.cpp src/test/unit/mcmc/hmc/hamiltonians/dense_e_metric_test.cpp src/test/unit/mcmc/hmc/hamiltonians/diag_e_metric_test.cpp src/test/unit/mcmc/hmc/hamiltonians/unit_e_metric_test.cpp src/test/unit/mcmc/hmc/mock_hmc.hpp src/test/unit/mcmc/sample_test.cpp src/test/unit/mcmc/test_csv_files/blocker.1.csv src/test/unit/mcmc/test_csv_files/blocker.2.csv src/test/unit/mcmc/test_csv_files/epil.1.csv src/test/unit/mcmc/test_csv_files/epil.2.csv src/test/unit/prob/distributions/multivariate/continuous/inv_wishart_test.cpp src/test/unit/prob/distributions/multivariate/discrete/multinomial_test.cpp src/test/unit/prob/welford_covar_estimator_test.cpp src/test/unit/prob/welford_var_estimator_test.cpp — Reply to this email directly or view it on GitHub.
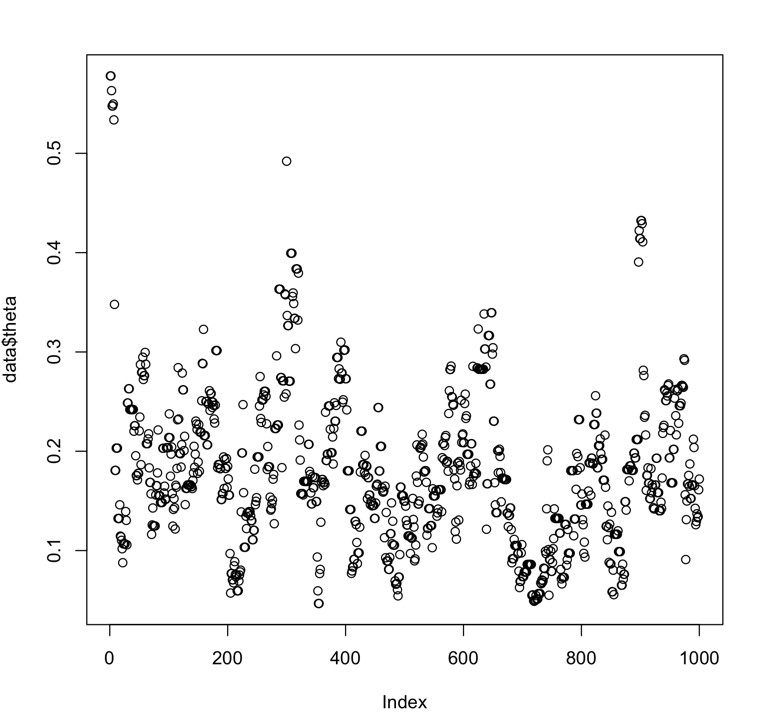
 . Right now it is always sets a=2 (which is also what emcee does). I'm not sure if it is worth tuning this or even varying it at all?
. Right now it is always sets a=2 (which is also what emcee does). I'm not sure if it is worth tuning this or even varying it at all?
Admit generic ensemble sampling within Stan's sampler engine.
Allows running multiple chains internally that can communicate with each other, possibly useful during adaptation. Allows ensemble samplers such as DREAM.