When dealing with Material Samples specifically using the Darwin Core Archive data format (i.e. star schema data structure used by GBIF/OBIS etc) I feel we have to make a decision between one of these options:
-
We continue to use an occurrence record (
rowType=Occurrence) for each record representing an item of preserved material. Issues with this approach relate to confusion (does the row represent the occurrence of the species in nature, or the material evidence of that, or some join across those or...) and to record identification (what isoccurrenceIDidentifying, noting that GBIF has enforced uniqueness constraints onoccurrenceID?).basisOfRecordis required here to distinguish "what" kind of occurrence the row is representing. -
We introduce a new type of row (
rowType=MaterialSample) and create a row for each material sample. When using this optionmaterialIDwould uniquely identify the row and basisOfRecord would be unnecessary. -
We promote more use of an event-oriented archive (
rowType=SamplingEvent) where the core of the star represents the gathering event or action of subsampling (e.g. extract of a leg) and an extension captures the material record. The extension could be an occurrence, which exists as a schema today or we create a new material one. This has issues, as you cannot today(!) have a third level of extension to capture multiple identifications of the specimen, or measurements, or other associated data with the material.
I think we need a decision on this to know what to do with basisOfRecord.
-
If we use rows of type
OccurrencethenbasisOfRecordis required to categorize the nature of the row (an observation or a specimen etc). We should then review if the current vocabulary is adequate or should be expanded. -
If we opt for rows that are of type
Material, then basisOfRecord can be ignored in this task group.
Does this make any sense to others, please?
I'd like to understand why it was made required in the IPT with a controlled vocabulary and if that provided the kinds of results people were expecting when that decision was made.
It is necessary for users to perform basic filtering on GBIF e.g. to exclude living specimens and fossils from a map. While not perfect it covered most of the basic categorization needs for search and reporting (removing ex-situ records is still an issue in some cases).
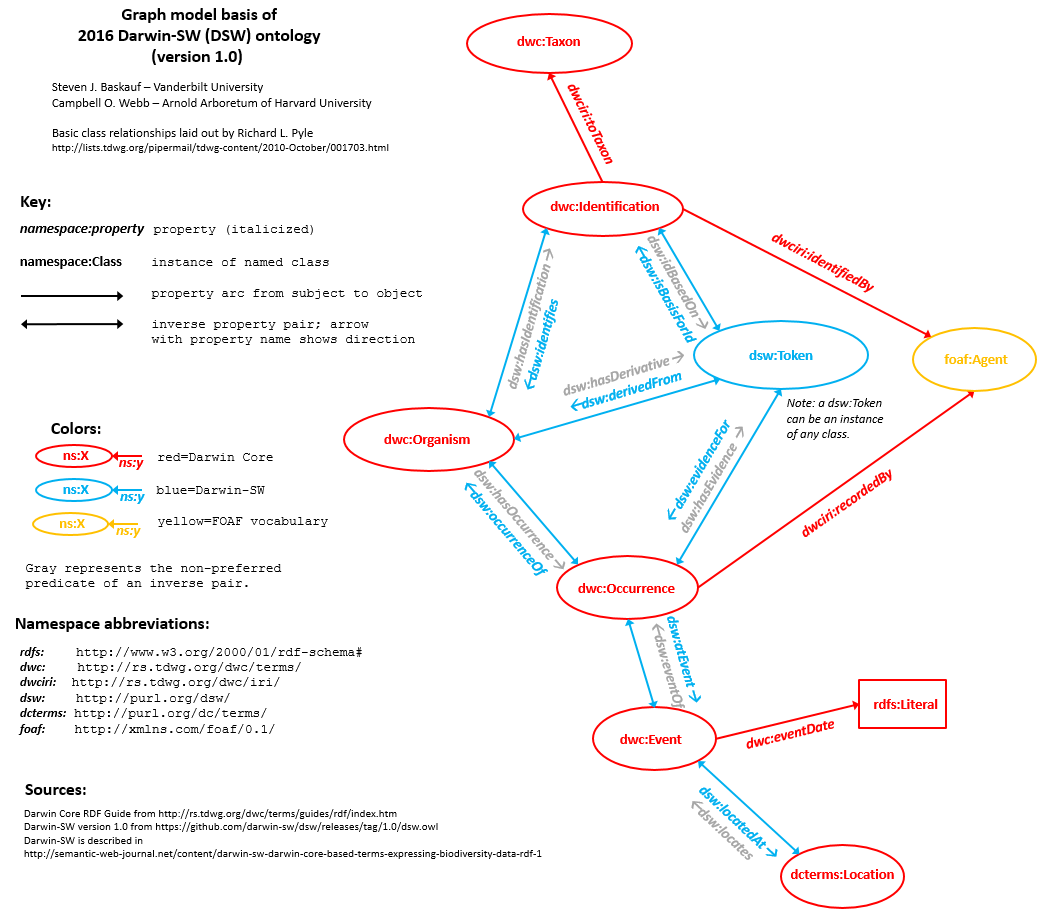

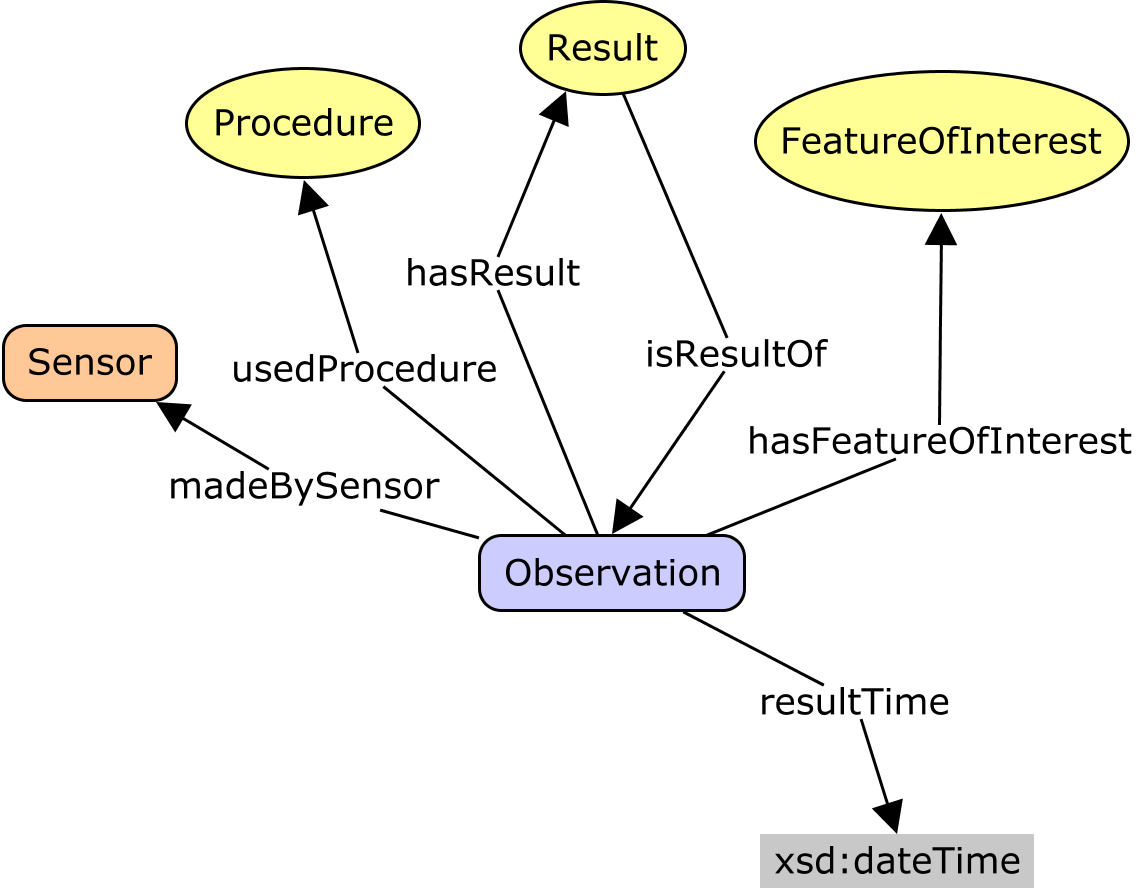
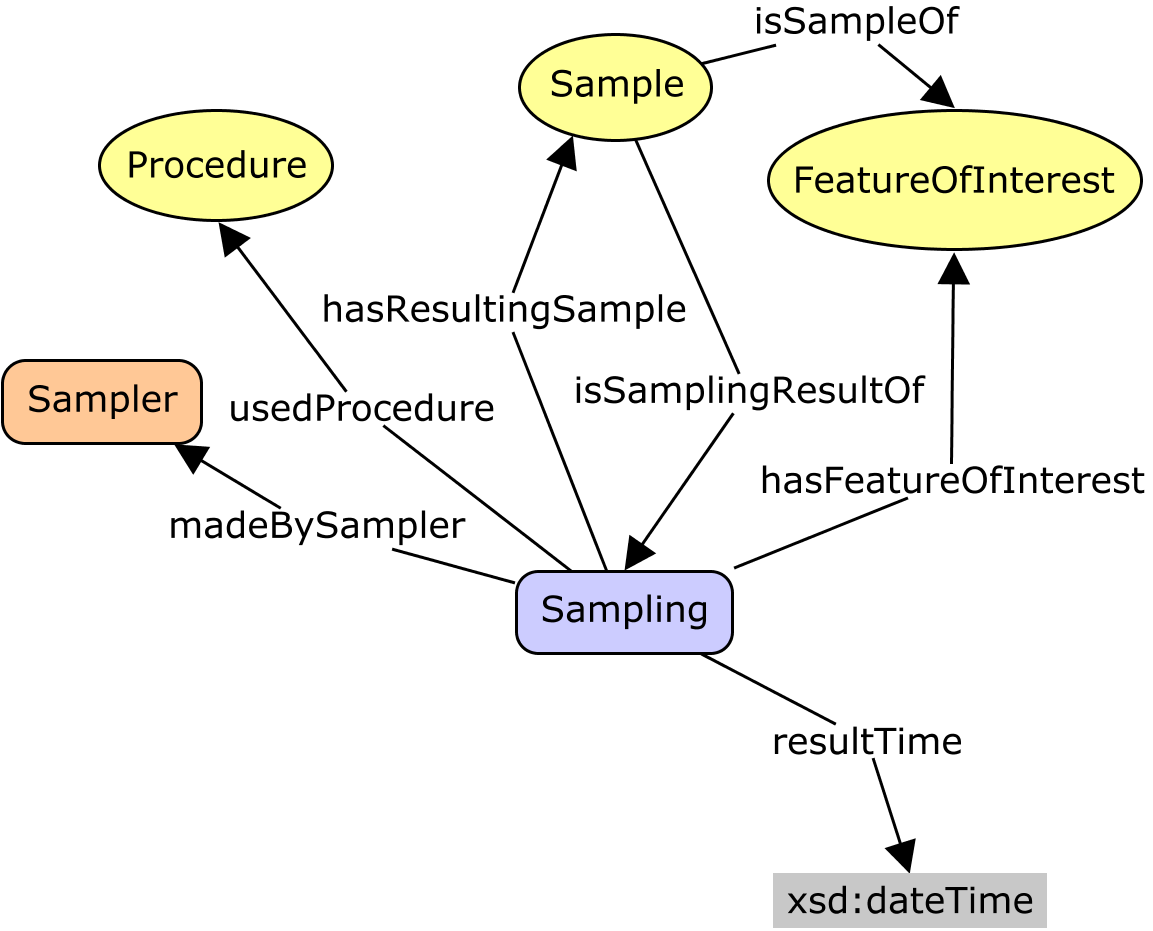

Current Darwin Core Placement/Definition
http://rs.tdwg.org/dwc/terms/basisOfRecord
this term is a property of Record-level
Defintion
The specific nature of the data record.
Examples
PreservedSpecimen, FossilSpecimen, LivingSpecimen, MaterialSample, Event, HumanObservation, MachineObservation, Taxon, Occurrence, MaterialCitation
Comments
Recommended best practice is to use the standard label of one of the Darwin Core classes.
See also
umbrella issue related to dwc:basisOfRecord and an Evidence class: https://github.com/tdwg/dwc/issues/302