could you explain this more clearly?
Open jrwin opened 7 years ago
could you explain this more clearly?
Please refer to #23
Hmm, the number of parameters remains the same, but the problem of DenseNets is that even though number of parameter remains the same, the connections determine performance..
I have seen that the performance of this model improves severely with the correct implementation of the Dense block (as @ahundt and I will attest to for the performance boost on segmentation tasks).
Unless I can execute the model with this change on the same dataset, and can observe noticeably improved performance, I don't want to merge this.
@jrwin Can you train a FCN model with and without this change on the same dataset to see how much the performance changed?
Also, @ahundt, is your dataset small enough to train within a few hours? Mine takes days, and I don't have access to the server I used before for the next 2 weeks, so I wont be able to run on my dataset.
@titu1994 , we have different use case. I try to do the liver and lesion segmentation for CT slice. And I trained a FCN with the modified densenet that can segment the liver pixels in CT slices with average DICE as 0.955. Previously, I used Unet achieve similar results. However, I tried to make multi-task segmentation with the densenetFCN(the modified version). The results is not good as Unet ever did. I am wondering how it happens. Then, I trained a densenetFCN with the 'x_up' to segment the liver, and it showed good as well. Currently, I am training a multi-tasks densenetFCN with 'x_up'.
These small modifications can definitely make a huge difference in performance! Could you clarify your wording here, did you mean to say:
I am training a multi-tasks densenetFCN without 'x_up'
In other words, you're running with this patch applied, correct? Looking at the large graph @titu1994 printed out in #23 It looks like you're removing one input from concatenate_101. (Could you print the layer name on your change to confirm?)
Here is a screenshot from the 100 layers paper:

And here is the screenshot from the original densenet paper:
 and the tiramisu author:
and the tiramisu author:
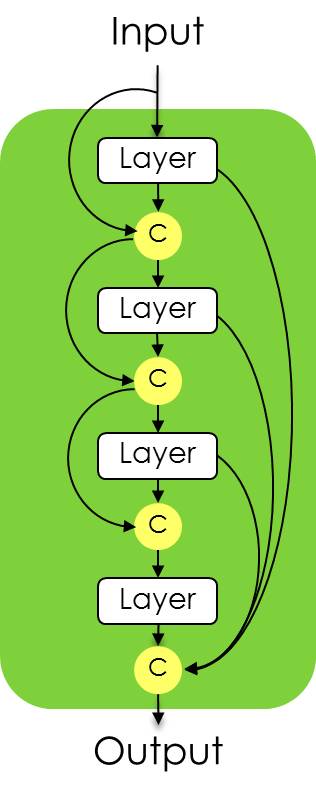 As you can see, a dense block concatenates everything to the output, including the input after the first "red" conv block is applied.
As you can see, a dense block concatenates everything to the output, including the input after the first "red" conv block is applied.
The correctness of the current implementation (without this pull request) proves to be the case even when comparing against the original author's published code, according to how I read it: https://github.com/SimJeg/FC-DenseNet/blob/master/FC-DenseNet.py#L108
Even with all of the above said, it is entirely conceivable that your change improves performance despite differing from what is implemented in the paper. :-)
Your PR here is certainly worth additional testing, but we would need some hard numbers on a standard dataset, see https://github.com/aurora95/Keras-FCN perhaps for code to train on pascal voc, and even if it does in fact improve results applying this change would probably need to be an optional flag, clearly labeling the flag as a deliberate departure from the paper. This is so people can still use a network equivalent to the original paper's implementation.
Ah wait now I'm a little mixed up, I have a meeting will look some more after
@titu1994 I looked into this further and you made this exact change in keras contrib :-) https://github.com/farizrahman4u/keras-contrib/commit/fe020327c4aa9325c6e0b98d4ee9ba7fc717bd14
After further review, I believe this PR is correct.
I would still like a performance test.
As far as I remember, I reverted that commit cause it diminished performance on the private dataset I was using.
In other words, you're running with this patch applied, correct?
@ahundt , it is right. But my densenetFCN is not the original one with 100 layers.
It was a simple one with 67 layers, where layers of each dense block in compression path is 3, 3, 4, 4, 6, init conv is 16, k is 12, compression rate is 0.5.
The following was the ploted model of 'exclude the input with the feature maps'. Lets call it NDoutput:

For comparison, the following was the ploted model of 'x_up'. Lets call it Doutput:

Base on my evaluation of the Liver segmentation on the LiTS dataset, the NDoutput get 0.955 Dice per case , and the Doutput get 0.935 Dice per case.
I am still confused now, for the Doutput combines more info(features) than the NDoutput, but Doutput got no better results on my evaluation.
exclude the input with the feature maps in the concatenation after the last desnblock