Thanks for your questions. For graph-level classification you essentially have two options:
- "hacky" version: you add a global node to the graph that is connected to all other nodes and run the standard protocol. You can then interpret the final value of this global node as graph-level label.
- global pooling: as a last layer, you can implement a global pooling layer that performs some form of pooling operation (optionally with attention mechanism as in https://arxiv.org/abs/1511.05493) over all graph nodes
For batch-wise training over multiple graph instances (of potentially different size) with an adjacency matrix each, you can feed them in the form of a block-diagonal adjacency matrix (each block corresponds to one graph instance) to the model, as illustrated in the figure below:
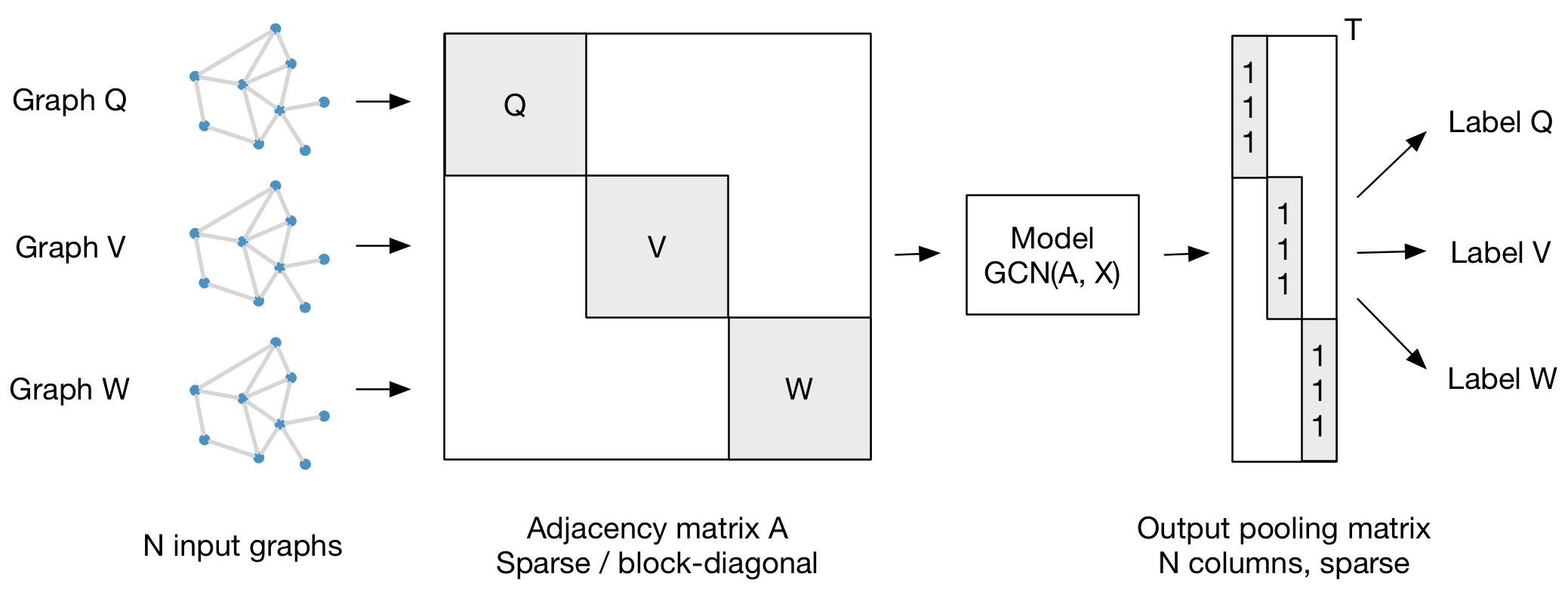
Hi I have two questions:
featureless=Falsewhen defining a newGraphConvolutionlayer. However, the loss is still computed for each node, and I was wondering how I should change your code.train.pyfor batch-wise training?Thanks for putting this together!