pileup format
- http://samtools.sourceforge.net/pileup.shtml
- https://en.wikipedia.org/wiki/Pileup_format
- </> (less-/greater-than sign) denotes a reference skip. This occurs, for example, if a base in the reference genome is intronic and a read maps to two flanking exons. If quality scores are given in a sixth column, they refer to the quality of the read and not the specific base.
- ^ (caret) marks the start of a read segment and the ASCII of the character following `^' minus 33 gives the mapping quality





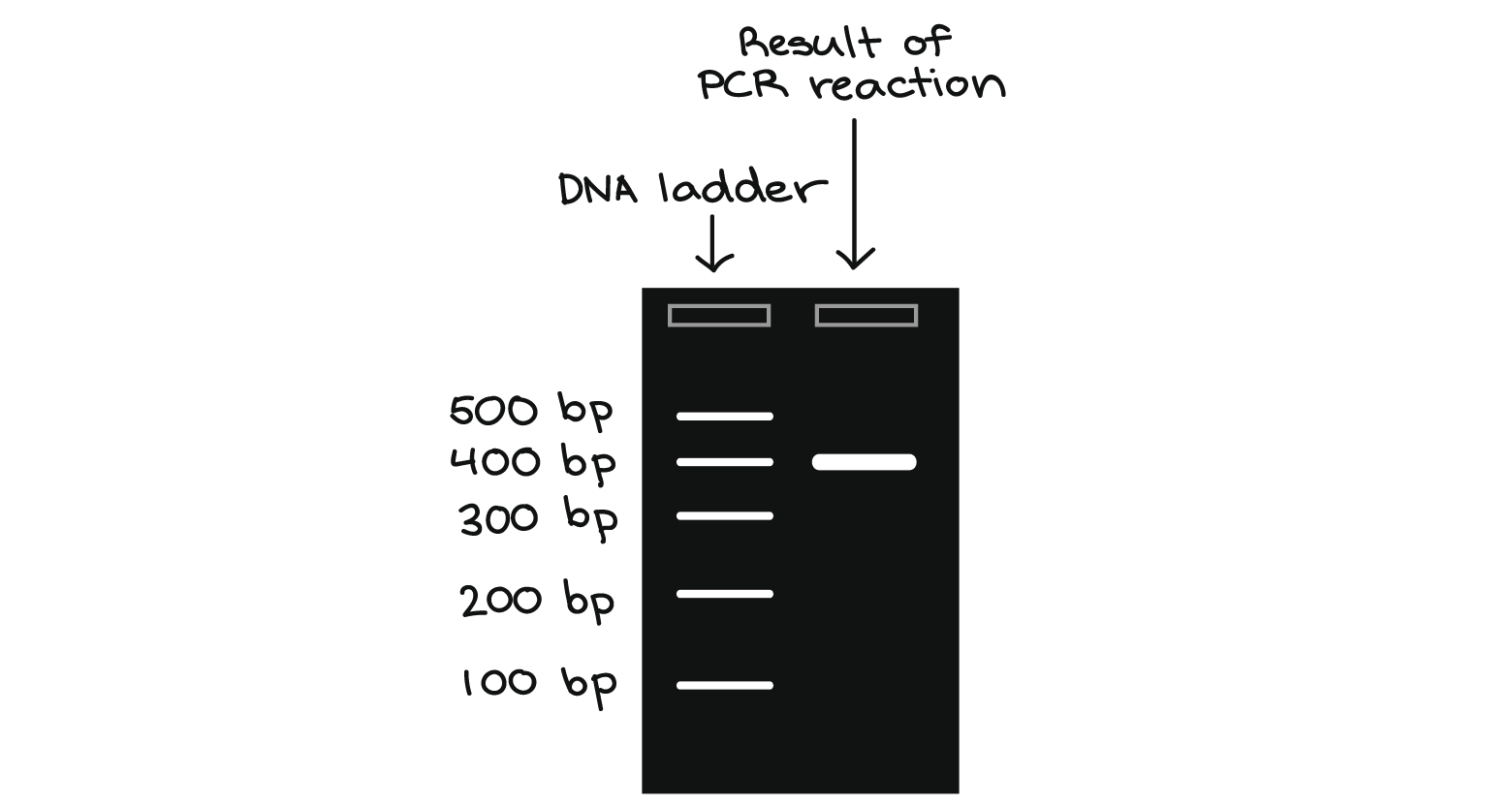
In addition to play with the source code, I also asked the Samtools community for help:
https://github.com/samtools/samtools/issues/1406 https://github.com/samtools/samtools/issues/1407 https://github.com/samtools/samtools/issues/1409