@wasserth HI sorry to disappoint, I'm am just an enthusiast amateur in medecine, I have no serious use for this software currently, still it is a potentially life saving advance in medecine. Noob question: I want to use the online version and give it as input a CT scan of a leg. So my question is: what are the supported formats for the CT scan single file? I know it must be a zip but once uncompressed, can it be a regular image (JPEG, PNG) or does the image needs to conform to a specific format? E.g can I input this? https://www.researchgate.net/figure/Patient-Abdominal-CT-scan-Normal-diameter-appendix-with-no-evidence-of-inflammation-of_fig2_323371468/amp


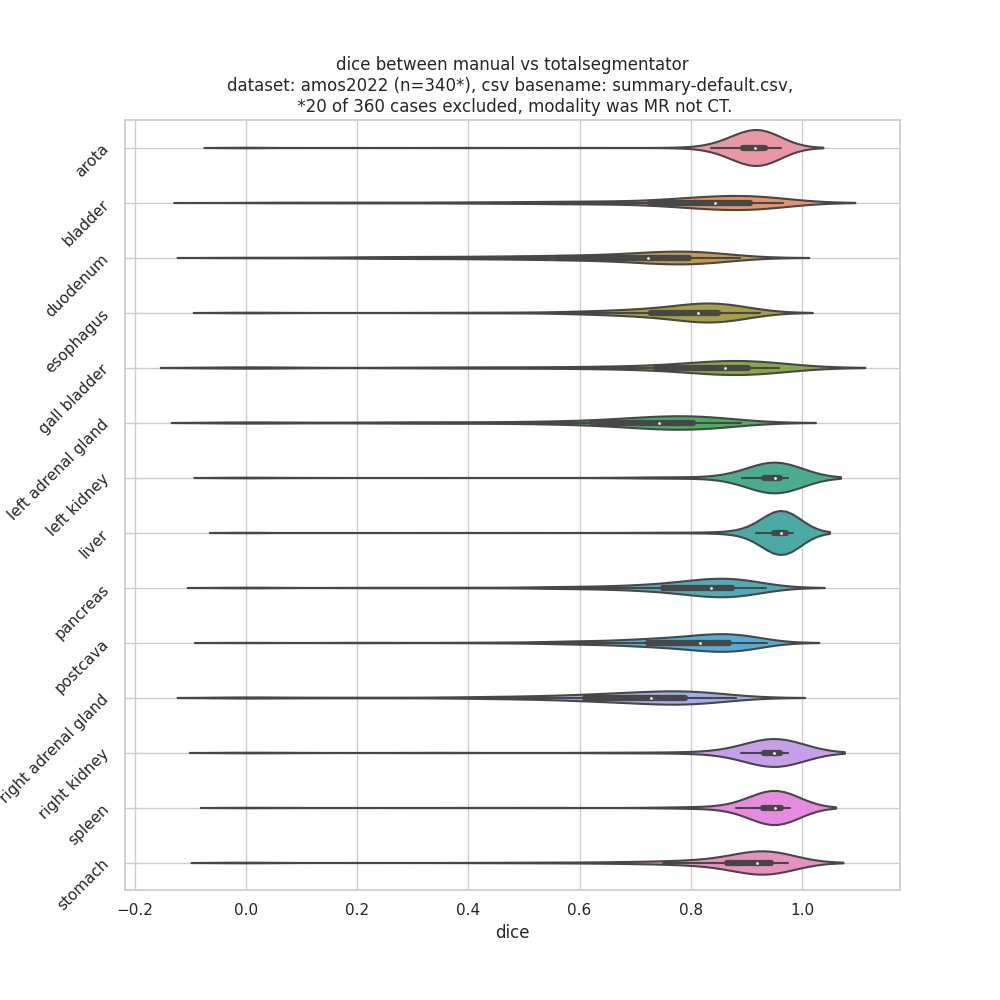
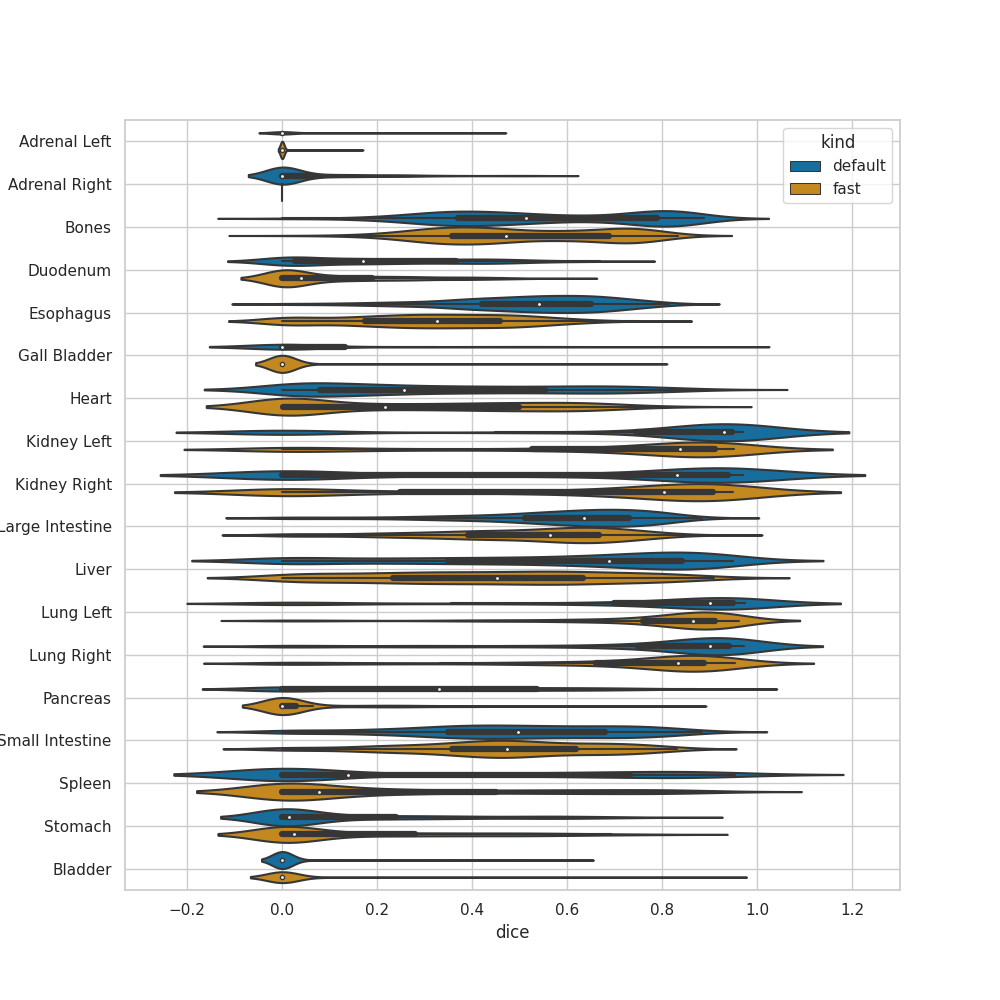
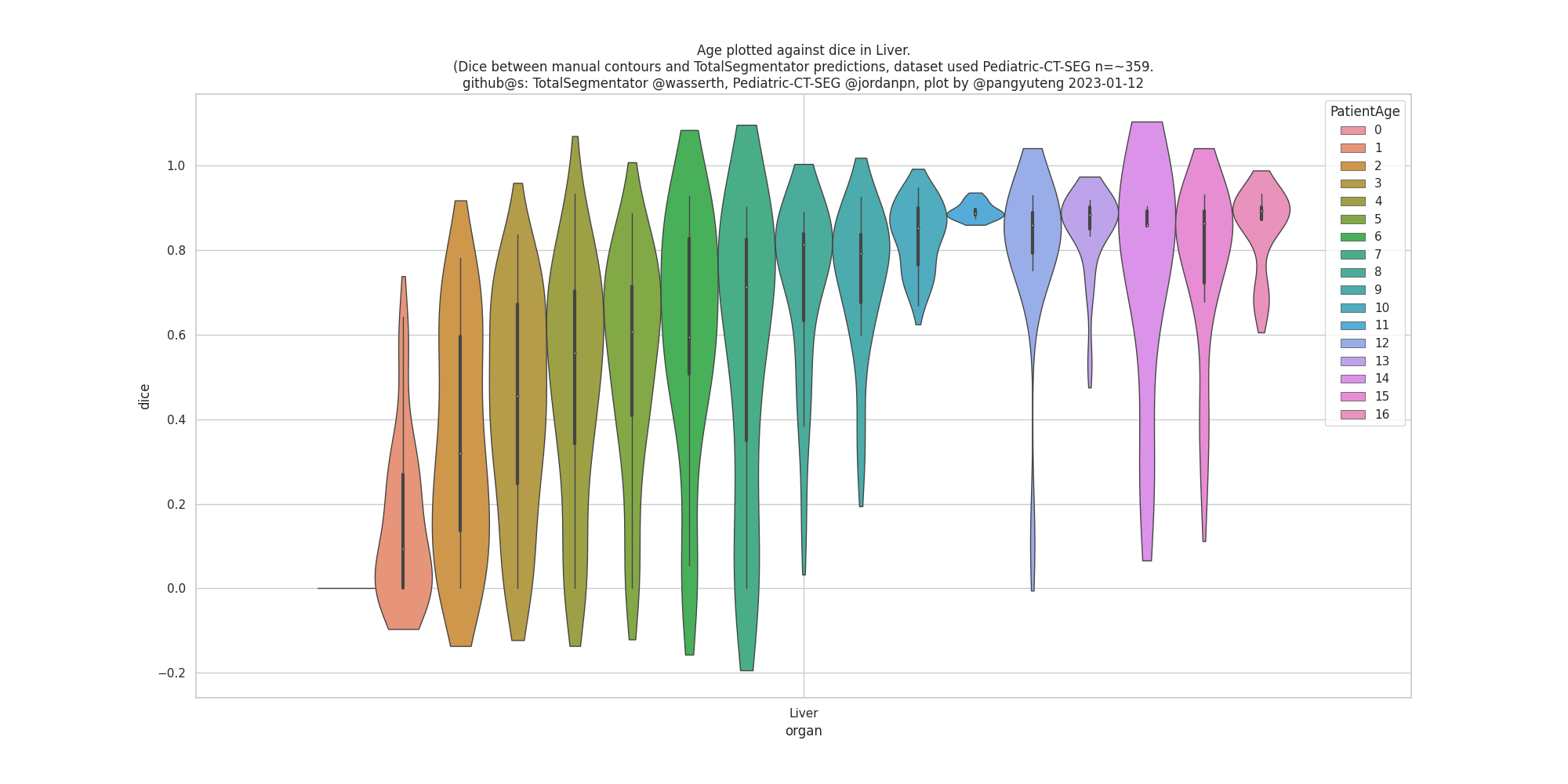
It would be really helpful for us to now who is using TotalSegmentator and what they are using it for. Therefore, we invite you to comment on this issue: What are you using TotalSegmentator for? What classes are you using? Are there any classes you are missing (we can try to add them in future releases)? Who are you (only if you want to disclose this)?