Hi,
Thanks for trying out our new version. Two things I can think of:
- By
readRDS(), to my experience, the object is indeed loaded into the environment but just not shown in the "environment" panel of your RStudio. If you try operators like:
obj <- readRDS("oldfile.rds")
obj@norm.dataIt should show you a list of normalized matrices. and similar for the obj@scale.data
- However, force doing those above should probably keep spamming error messages on your screen. You can downgrade the package back to version 1.0.1 and normally load it, and have checks on the colnames and rownames of the matrices. In rliger2.0.0, we added strict checkpoints for cell and feature consistency to prevent potentially erroneous operations. What you will want to look at is, regard the row/colnames of the
obj@raw.dataas the baseline, see if those innorm.dataare identical.scale.datashould be feature subset and transposed (cell x gene) in the old version, and you can see if the features are available inrownames(obj@raw.data[["datasetName"]]).
Above is kind of the overall idea of what should be taken care of, please feel free to reach out if you still see a problem.
Here's a figure on the new documentation website that roughly shows how the structure of a liger object is refactored:
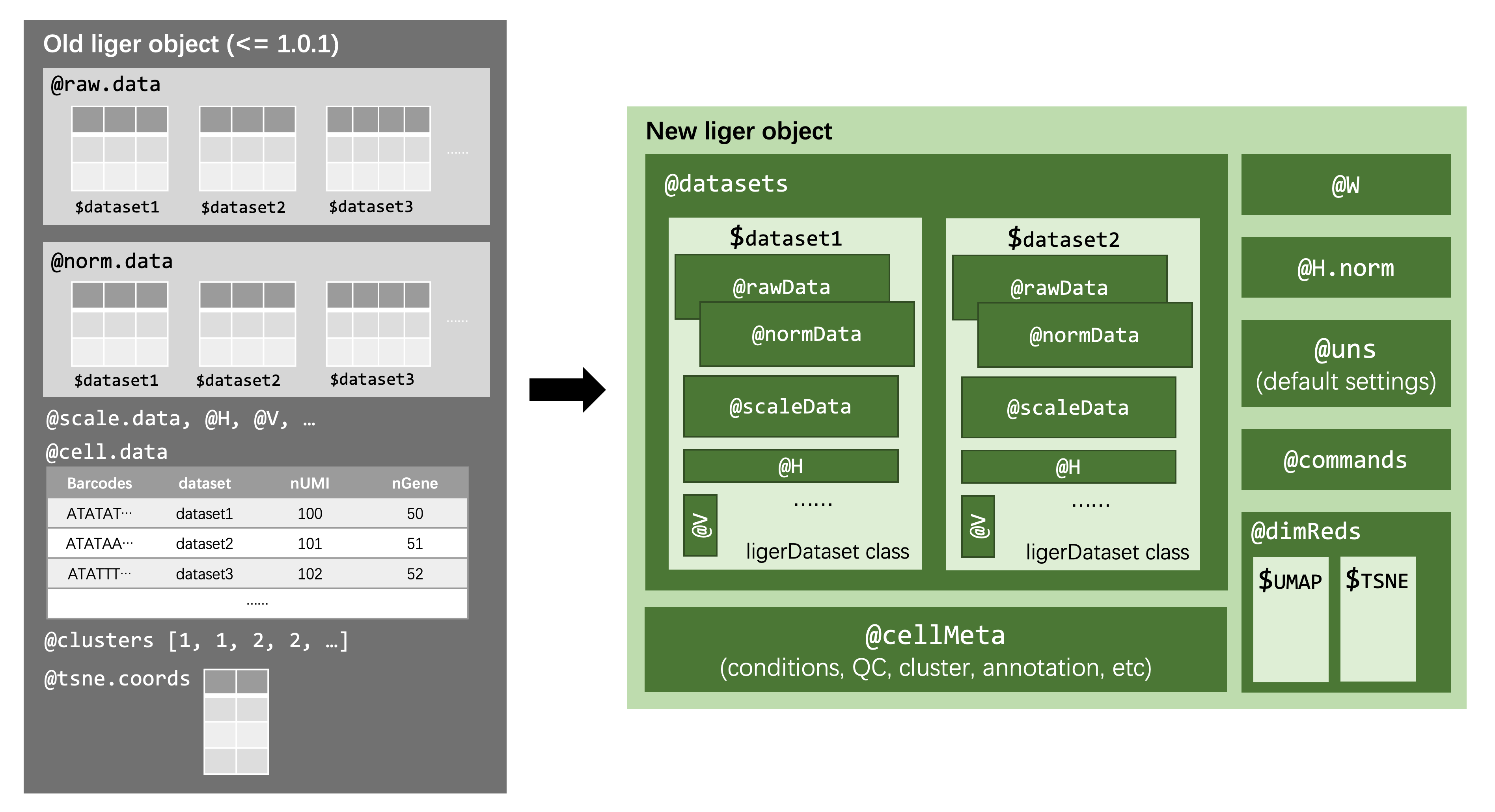
Let's keep this Issue open for people coming for rliger2 structure issues.
Appreciate it! Yichen
Dear Welch lab/liger team,
I'm hoping for some help with figuring out how to load in an old liger object that is giving me issues.
The object is from a previously published dataset from early 2023. I'm happy to include a link to the dataset here, if necessary.
When I first attempt to load in the object using readRDS I get the following error:
Error in .checkObjVersion(): ! Old version of liger object is detected. Please run: object <- convertOldLiger(object)When I then try to run the indicated convertOldLiger function, I get the following error:
! Skipped slot U which is not available. ✖ Inconsistent row ID in slot normData. Error in validObject(.Object) : invalid class “ligerDataset” object: Features in scaleData not found from dataset: [features]and the object fails to load.
The problem is, because I can't get the object loaded in any form, I can't try and troubleshoot any individual steps of the object conversion function.
Any tips, advice, or feedback would be greatly appreciated! Thank you very much for your time.