Except for a small typo, I think your example should do the correct thing. However, if you want to use redundant arrays as input, you might want to check that they have been actually properly symmetrized. E.g. one could use a function like
from wilson.util import smeftutil
def C_arrays_to_C_wcxf(C_arrays):
C_wcxf = smeftutil.arrays2wcxf_nonred(smeftutil.add_missing(C_arrays))
C_arr_sym = smeftutil.wcxf2arrays_symmetrized(C_wcxf)
for k,v in C_arrays.items():
if not np.all(C_arr_sym[k] == v):
raise ValueError(f'Input array "{k}" is not properly symmetrized.')
return C_wcxfWe could think about including such a function in a Wilson.from_C_arrays class method that would work similar to Wilson.from_wc. We just might have to find a better name :wink:
Another idea would be to check whether the input dictionary contains keys for redundant arrays (i.e. lq1, lq3, etc.) or for non-redundant entries (i.e. lq1_1111, lq1_1112, etc.). Then both options could be accepted as inputs and for the first option one could just call C_arrays_to_C_wcxf internally.
When matching from a UV theory onto the SMEFT, often one gets pretty simple generic formulas for the SMEFT coefficients. As an example, take this from the wilson paper (bottom of page 9) But actually typing out all the coefficients is tedious and error prone. Again from there, you give the example code
But actually typing out all the coefficients is tedious and error prone. Again from there, you give the example code
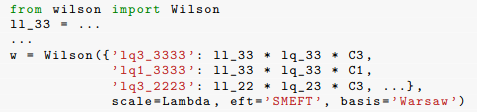 If I'm correct, there are actually another 17 coefficients hidden in that "..." that you didn't bother to type out, and of course you have to remember which are the non-redundant ones.
If I'm correct, there are actually another 17 coefficients hidden in that "..." that you didn't bother to type out, and of course you have to remember which are the non-redundant ones.
Instead, it's pretty easy to use the following code
to generate all the wilson coefficients (in what should be the basis where coefficients have the same symmetries as the operators). Then you can do
to get a dictionary with just the non-redundant coefficients needed to initialise a Wilson instance.
My questions are: 1) Does that combination of functions do the correct thing? I.e. take care of the symmetries correctly etc. I talked to @peterstangl last week and he agreed that there was some internal function that does this, and I believe
arrays2wcxf_nonredis what I want, notarrays2wcxf. and 2) Do you think this has wider use, and if so, what about making this an official way to get aWilsoninstance? I would say using the numpy summation makes it much easier to see if you have the correct formula, and much less typo prone.