Here is a quick guide for you to start with. This is a good idea for making an actual sample later on.
Create your 2D binary mask. Use a mask with ones and nans. If you have it as ones and zeroes do something like this:
mask= np.nan*np.ones_like(image_2d)
Extract x y z images from the point cloud. Check https://github.com/zivid/python-samples/blob/master/read_zdf.py but extract x y z as separate images, i.e.
x = np.dstack([point_cloud["x"]]) y = np.dstack([point_cloud["y"]]) z = np.dstack([point_cloud["z"]])
Then comes the masking. You can use numpy.multiply() to multiply each of the x y z images with a mask. That's pixel-wise multiplication.
x_new = np.multiply(x,mask) y_new = np.multiply(y,mask) z_new = np.multiply(z,mask)
Create the point cloud from the resulting x y z images.
xyz_new = np.dstack([x_new, y_new, z_new]) pc_new = np.dstack([xyz_new, image])
This is the idea. Please let us know if it works for you.

Hello! I am looking for some good method to mask an existing pointcloud obtained using Zivid.
I have the pointcloud (PLY and ZDF)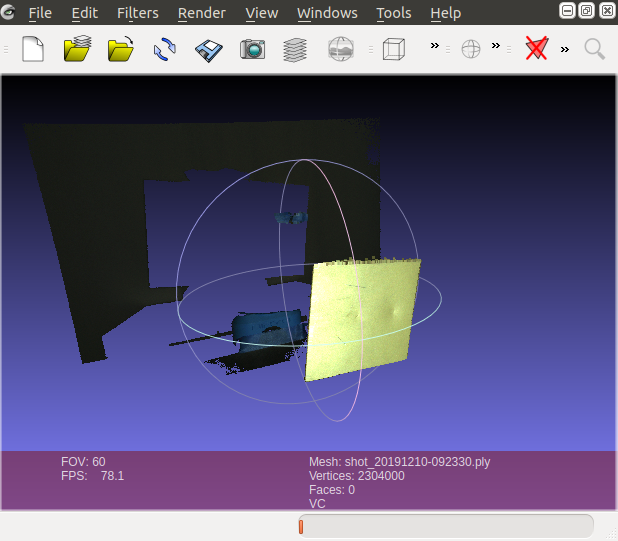
I have the RGB image: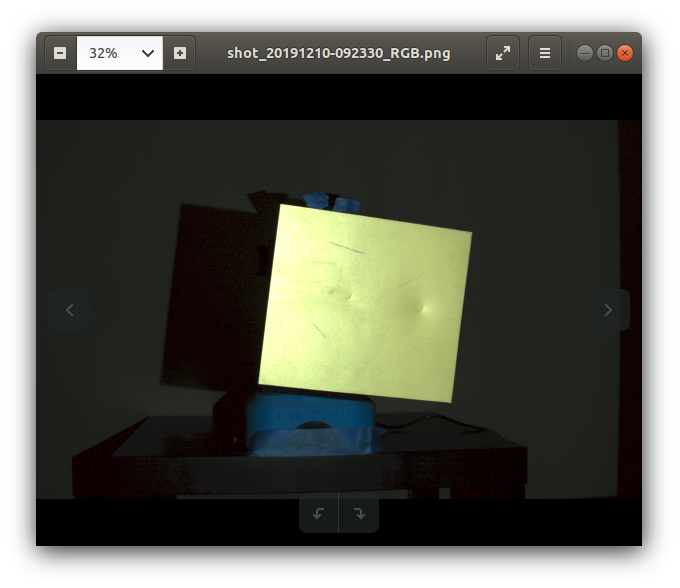
I have a binary mask that isolates the yellow plate on the RGB image. I also have a cv2 contour based upon this binary mask.
The point cloud and the RGB are zivid based so they have the same number of pixels. The question is regarding how I could use the binary mask to remove all the xyz points masked by the black picture.
Any ideas welcome.
thanks!
@pbermell