Abnormality-Detection-in-Mammography
Motivation
When I was looking for a topic for the capstone project in the data science bootcamp program, my wife had her first time mammogrpahy. As the initial diagnose result was ambiguous, she had to make a couple of follow-up visits. Later, I got to know that she felt a lot of pain during the procedure. I naturally got interested in mammography diagnose and found that computer vision and deep learing could be an alternative way to read and decide the mammogrpy result.
Background
Breast cancer is the second leading cause of deaths among American women. The average risk of a woman in the United States developing breast cancer sometime in her life is approximately 12.4% 1. Screen x-ray mammography have been adopted worldwide to help detect cancer in its early stages. As a result, we've seen a 20-40% mortality reduction [2]. In recent years, the prevalence of digital mammogram images have made it possible to apply deep learning methods to cancer detection [3]. Advances in deep neural networks enable automatic learning from large-scale image data sets and detecting abnormalities in mammography [4, 5].
Considering the benefits of using deep learning in image classification problem (e.g., automatic feature extraction from raw data), I developed a deep Convolutional Neural Network (CNN) that is trained to read mammography images and classify them into the following five instances:
- Normal
- Benign Calcification
- Benign Mass
- Malignant Calcification
- Malignant Mass
Data Source
I obtained mammography images from the DDSM and CBIS-DDSM databases. The DDSM (Digital Database of Screening Mammography) is a database of 2,620 scanned film mammography studies. It contains normal, benign, and malignant cases with verified pathology information. The CBIS-DDSM (Curated Breast Imaging Subset of DDSM) is a subset of the DDSM database curated by a trained mammographer.
Notebooks
- CBIS-DDSM file rename and processing -> CBIS-DDSM_Rename_DICOM_Files.ipynb
- CBIS-DDSM patch extraction -> [CBIS-DDSM Patch_Extraction_256x256.ipynb](CBIS-DDSM Patch_Extraction_256x256.ipynb)
- DDSM file format conversion -> [lJPEG to PNG Conversion.ipynb](lJPEG to PNG Conversion.ipynb)
- DDSM data processing -> DDSM_Patch_Extraction_256x256.ipynb
- Data Labeling -> Data_Labeling.ipynb
- Data Preparation for model training -> Data_Preparation_for_CNN.ipynb
- CNN training -> [CNN Development and Evaluation.ipynb](CNN Development and Evaluation.ipynb)
- Utility functions (python file) ->img_processing_256.py
Artifacts Removal
The raw mammography images contain artifacts which could be a major issue in the CNN development. To remove the artifacts, I created a mask image as shown below for each raw image by selecting the largest object from a binary image and filled white gaps (i.e., artifacts) in the background image. I used the Otsu segmentation method to differentiate the breast image area with the background image area for the artifacts removal. Then, the boundary of the breast image was smoothed using the openCv morphologyEx method

Patch Extraction
Considering the size of data sets and available computing power, I decided to develop a patch classifier rather than a whole image classifier. For this purpose, image patch extractions for the normal and abnormal images were conducted in two different way:
- Patches for the normal images were randomly extracted from within the breast image area
- Patches for the abnormal images were created by sampling from the center and around the center of ROI area

CNN Models
I designed a baseline model with a VGG (Visual Geometry Group) type structure, which includes a block of two convolutional layers with small 3×3 filters followed by a max pooling layer. The final model has four repeated blocks, and each block has a batch normalization layer followed by a max pooling layer and dropout layer. Each convolutional layer has 3×3 filters, ReLU activation, and he_uniform kernel initializer with same padding, ensuring the output feature maps have the same width and height. The architecture of the developed CNN is shown below.

You can also download the trained CNN models
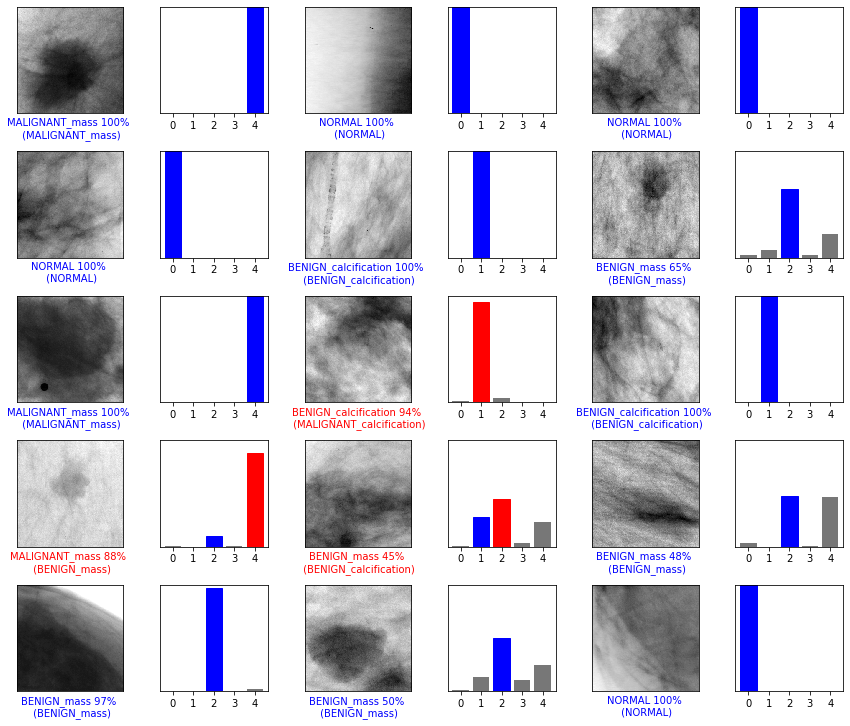
Examples of Predictions
Figure below shows that correct prediction labels are blue and incorrect prediction labels are red. The number gives the percentage for the predicted label.
Conclusions and Future Studies
-
The achieved accuracy of the multi-class classification model was 90.7%, but the accuracy is not a proper performance measure under the unbalanced data condition.
-
The results of precision and recall for the abnormal classes (e.g., Benign Calcification, Benign Mass, Malignant Calcification, and Malignant Mass) in the multi-class classification model were relatively lower than the estimated accuracy. The recall value for each abnormal class was 68.4%, 50.5%, 35.8%, and 47.1%, respectively, while the precision value was 68.8%, 48.5%, 56.7%, and 57.1%, respectively. However, the weighted average of the precision and the weighted average of recall were 89.8% and 90.7%, respectively.
-
The precision and recall values for detecting abnormalities (e.g., binary classification) were 98.4% and 89.2%.
-
This project will be enhanced by investigating the ways to increase the precision and recall values of the multi-class classification model. An immediate extension of this project is to investigate the model performance after adding additional blocks/layers into the existing CNN model and tuning hyper-parameters. In the meantime, I will examine the data imbalance issue with both over-sampling and under-sampling techniques. Additionally, I will improve the developed CNN model by integrating with a whole image classifier.
References
-
Lotter, William, et al. "Robust breast cancer detection in mammography and digital breast tomosynthesis using annotation-efficient deep learning approach." arXiv preprint arXiv:1912.11027 (2019).
-
Abdelhafiz, Dina, et al. "Deep convolutional neural networks for mammography: advances, challenges and applications." BMC bioinformatics 20.11 (2019): 281.
-
Nelson, Heidi D., et al. "Factors associated with rates of false-positive and false-negative results from digital mammography screening: an analysis of registry data." Annals of internal medicine 164.4 (2016): 226-235.
-
Xi, Pengcheng, Chang Shu, and Rafik Goubran. "Abnormality detection in mammography using deep convolutional neural networks." 2018 IEEE International Symposium on Medical Measurements and Applications (MeMeA). IEEE, 2018.
-
Rebecca Sawyer Lee, Francisco Gimenez, Assaf Hoogi , Daniel Rubin (2016). Curated Breast Imaging Subset of DDSM [Dataset]. The Cancer Imaging Archive. DOI: 10.7937/K9/TCIA.2016.7O02S9CY https://github.com/trane293/DDSMUtility
-
Lehman, Constance D., et al. "National performance benchmarks for modern screening digital mammography: update from the Breast Cancer Surveillance Consortium." Radiology 283.1 (2017): 49-58. Shen, Li, et al. "Deep learning to improve breast cancer detection on screening mammography." Scientific reports 9.1 (2019): 1-12.