No Bullshit Converter is a tool that helps you translate Pytorch models into Keras/Tensorflow/TFLite graphs without losing your mind.
- Supports a wide range of architectures
- [x] Control flow ops (If, For, While)
- [x] Recurrent layers (LSTM, GRU)
- [x] Stateful modules
- [x] Arbitrary torch functions
- Simple
- Flexible
- Efficient
- Sanity-preserving, with clear mistake messaging
[!IMPORTANT]
Nobuco only supports Keras 2 at the moment. If you'd like to use the new multi-backend Keras 3, please bump up the related issue: https://github.com/keras-team/keras/issues/19314
Installation 

pip install -U nobucoTable of Contents
- Essentials
- Channel order wizardry
- Implementation mismatch: pick your poison
- Going dynamic
- No, that's too dynamic!
- In-place operations
- A little white lie: tracing mode
- Ad hoc modifications
- So we put a converter inside your converter
- But was it worth it?
- Nobuco knowledge base
Essentials
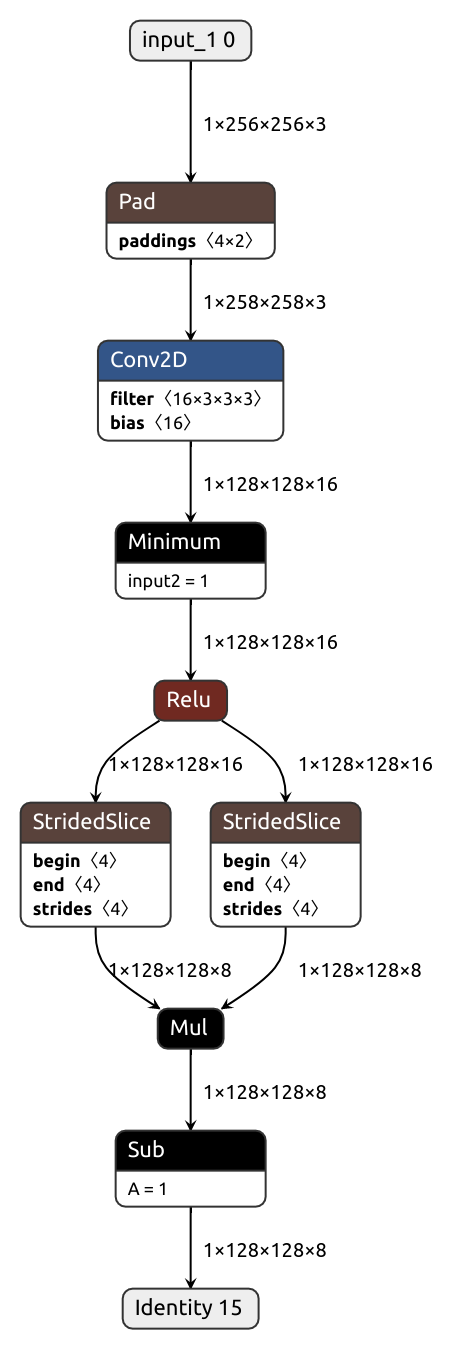
Suppose we want to convert a Pytorch module similar to this one:
class MyModule(nn.Module):
def __init__(self):
super().__init__()
self.conv = nn.Conv2d(3, 16, kernel_size=(3, 3), padding=(1, 1), stride=(2, 2))
def forward(self, x):
x = self.conv(x)
x = nn.Hardsigmoid()(x)
x = 1 - x[:, ::2] * x[:, 1::2]
return xThe process is exactly what you would expect. Instantiate the module, create dummy inputs and call the magic function:
import nobuco
from nobuco import ChannelOrder, ChannelOrderingStrategy
from nobuco.layers.weight import WeightLayerdummy_image = torch.rand(size=(1, 3, 256, 256))
pytorch_module = MyModule().eval()
keras_model = nobuco.pytorch_to_keras(
pytorch_module,
args=[dummy_image], kwargs=None,
inputs_channel_order=ChannelOrder.TENSORFLOW,
outputs_channel_order=ChannelOrder.TENSORFLOW
)Aaaand done! That's all it takes to... hold on, what's that?
Nobuco says it doesn't know how to handle hard sigmoid.
Apparently, it's our job to provide a node converter for either F.hardsigmoid or the enclosing Hardsigmoid module (or the entire MyModule, but that makes little sense). Here, we'll go for the former.
Conversion is done directly. No layers upon layers of abstraction, no obscure intermediate representation.
A node converter is just a Callable that takes the same arguments as the corresponding node in Pytorch and outputs an equivalent node in Tensorflow.
The converted node preserves the original node's signature, but Pytorch tensors replaced with Tensorflow counterparts (be that tf.Tensor, KerasTensor, tf.Variable, or ResourceVariable).
This should do the trick:
@nobuco.converter(F.hardsigmoid, channel_ordering_strategy=ChannelOrderingStrategy.MINIMUM_TRANSPOSITIONS)
def hardsigmoid(input: torch.Tensor, inplace: bool = False):
return lambda input, inplace=False: tf.keras.activations.hard_sigmoid(input)It works, but the outputs don't quite match. Perhaps we should check how Pytorch and Tensorflow define hard sigmoid. And sure enough, their implementations differ. Have to type in the formula manually, I guess...
@nobuco.converter(F.hardsigmoid, channel_ordering_strategy=ChannelOrderingStrategy.MINIMUM_TRANSPOSITIONS)
def hardsigmoid(input: torch.Tensor, inplace: bool = False):
return lambda input, inplace=False: tf.clip_by_value(input/6 + 1/2, clip_value_min=0, clip_value_max=1)And the happy result:
The example above is artificial, but it illustrates the point. It's not feasible to provide a node converter for every existing Pytorch op. There are literally thousands of them! Best we can do without the converter constantly lacking essential functionality, being riddled with bugs, doing weird stuff and breaking apart with every other PT/TF release is to keep the tool simple and customizable, make it clear where a problem comes from and let the user sort things out. Usually it's easy for a human to translate an isolated operation from one framework to another. Reproducing the graph structure is a different matter entirely. For that, Nobuco has you covered!
Nobuco lets you intervene in conversion at each step, asks for help where needed and doesn't bother you with routine stuff.
https://user-images.githubusercontent.com/2457934/233740603-cc11acc5-cd6b-48c8-b089-ff3ead772dd0.mp4
With an IDE, you can jump right where the node was [I]nvoked, [D]efined and [C]onverted
Channel order wizardry
Some operations assume its input tensors have a channel dimension. And as you probably know, Pytorch and Tensorflow do not agree on the layout of such tensors. Pytorch adopts channel-first layout (BCH, BCHW, etc.) while Tensorflow works efficiently with channel-last tensors (BHC, BHWC, ...). Transposing tensors between the two layouts incurs non-trivial overhead as generally, tensor data must be physically rearranged. In an effort to keep that overhead to the minimum, Nobuco does layout coercions lazily. A couple of things are needed to make it possible:
- Tensorflow tensors are augmented with an additional property which stores their channel order, either Pytorch (channel first) or Tensorflow (channel last) style.
- Node converters have requirements on what channel order their inputs must have. Said requirements are expressed with
channel_ordering_strategyargument.
Channel ordering strategies are
FORCE_TENSORFLOW_ORDER- Input tensors will be coerced to Tensorflow channel order.
- Convenient for converting channel-aware operations (convolution, batchnorm).
FORCE_PYTORCH_ORDER- Input tensors entering the node will look exactly as they do in the original Pytorch graph.
- Use it when the node does not interpret its input tensors as having a channel dimension (linear, matmul).
MINIMUM_TRANSPOSITIONS- The channel order is decided by a majority vote (whichever prevails among the inputs). This way the number of coercions (i.e. tensor transpositions) is kept to the minimum. It also means whenever there's only one input, it will be left untouched.
- Best choice for element-wise ops (most activations).
MANUAL- You are on your own. In exchange for unrestricted freedom, you take responsibility to coerce input tensors to suitable channel order and to also annotate output tensors with their order.
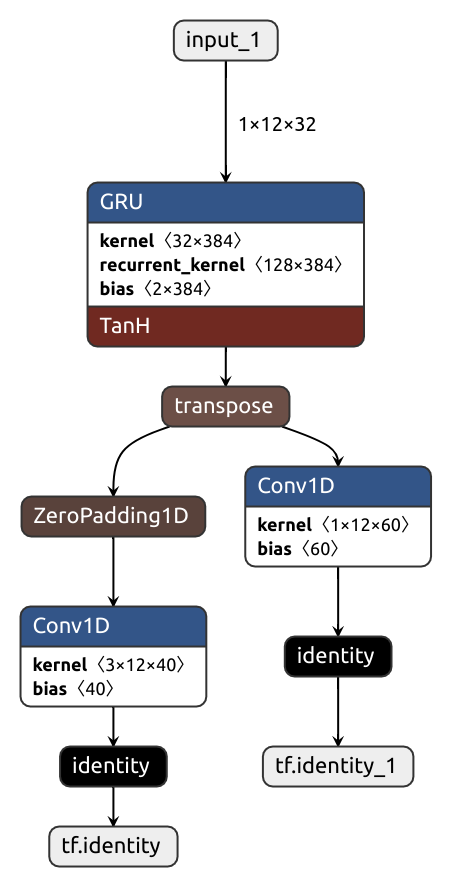
The simple lazy approach makes wonders in most situations, but sometimes it produces suboptimal graphs. Consider the code below. Imagine this is some sort of text processing network. It first applies a GRU layer which assumes the inputs do not have a channel dimension, so its input/output layouts are the same in both Pytorch and Tensorflow. But then, the outputs are passed to a couple of 1D convolutions which are channel-aware. Because of that, a transpose op must be put in the converted graph.
class MyModule(nn.Module):
def __init__(self):
super().__init__()
self.gru = nn.GRU(32, 128, num_layers=1, batch_first=True, bidirectional=False)
self.conv1 = nn.Conv1d(12, 40, kernel_size=3, padding=1)
self.conv2 = nn.Conv1d(12, 60, kernel_size=1, padding=0)
def forward(self, x):
x, hx = self.gru(x)
x1 = self.conv1(x)
x2 = self.conv2(x)
return x1, x2
pytorch_module = MyModule().eval()
inputs = [
torch.normal(0, 1, size=(1, 12, 32)),
]
keras_model = nobuco.pytorch_to_keras(
pytorch_module, inputs,
inputs_channel_order=ChannelOrder.PYTORCH,
)The laziness shoots us in the foot here, and we get not one transpose but two:
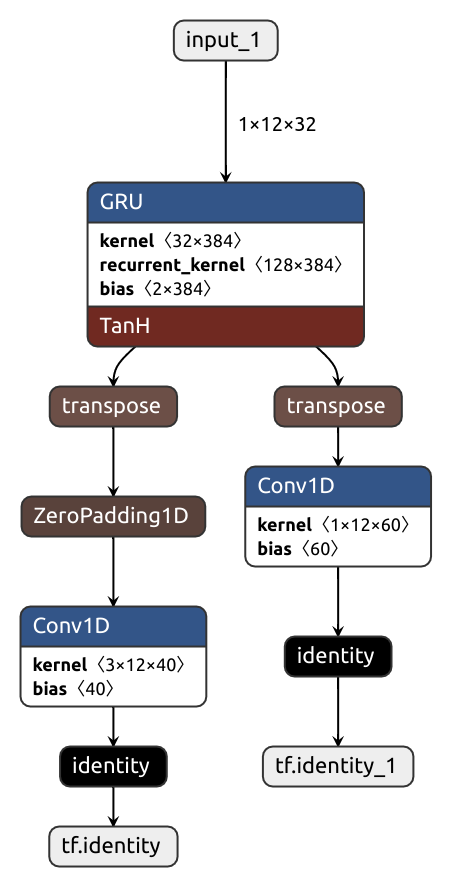
For such occasions, there's two brethren functions: force_tensorflow_order and force_pytorch_order.
x, hx = self.gru(x)
x = nobuco.force_tensorflow_order(x)
x1 = self.conv1(x)
x2 = self.conv2(x)
In case you are curious, the implementation is trivial:
@nobuco.traceable
def force_tensorflow_order(inputs):
return inputs
@nobuco.converter(force_tensorflow_order, channel_ordering_strategy=ChannelOrderingStrategy.FORCE_TENSORFLOW_ORDER)
def converter_force_tensorflow_order(inputs):
return lambda inputs: inputsforce_pytorch_order is defined analogously.
Implementation mismatch: pick your poison
Sometimes, Pytorch and Tensorflow just don't go along.
In case of hardsigmoid, it's a mere inconvenience, but it can be much more sinister.
Take the model below, for example.
class MyModule(nn.Module):
def __init__(self):
super().__init__()
self.factor = 4
self.conv = nn.Conv2d(3*self.factor**2, 3*self.factor**2, kernel_size=1)
def forward(self, x):
x = nn.PixelUnshuffle(self.factor)(x)
x = self.conv(x)
x = nn.PixelShuffle(self.factor)(x)
return xIdeally, there would only be three nodes in the converted graph. That's not what we get, though.
Tensorflow does not have pixel_unshuffle/pixel_shuffle.
Their closest counterparts, tf.nn.space_to_depth/tf.nn.depth_to_space,
do almost the same thing but not quite: output channels are in a different order.
The order must be fixed with a pricey transpose, no way around that. Or is there?
![]()
Instead of emulating an absent Pytorch op in Tensorflow, we might do the procedure in reverse: provide a Pytorch implementation for the Tensorflow node we want to convert to. The overhead would be carried by the original Pytorch model leaving the converted graph nice and clean.
from nobuco.addons.torch.space_to_depth import SpaceToDepth
from nobuco.addons.torch.depth_to_space import DepthToSpace
class MyModuleTFOptimized(nn.Module):
def __init__(self):
super().__init__()
self.factor = 4
self.conv = nn.Conv2d(3*self.factor**2, 3*self.factor**2, kernel_size=1)
def forward(self, x):
x = SpaceToDepth(self.factor)(x)
x = self.conv(x)
x = DepthToSpace(self.factor)(x)
return x
![]()
| Torch-optimized | Tensorflow-optimized |
|---|---|
| **Torch implementation** ```python F.pixel_unshuffle ``` **Tensorflow converter** ```python @nobuco.converter(F.pixel_unshuffle, channel_ordering_strategy=ChannelOrderingStrategy.FORCE_TENSORFLOW_ORDER) def converter_pixel_unshuffle(input: Tensor, downscale_factor: _int): def func(input, downscale_factor): x = tf.nn.space_to_depth(input, downscale_factor) x = channel_interleave2d(x, downscale_factor, reverse=True) return x return func def channel_interleave2d(x, block_size: int, reverse: bool): b, h, w, c = x.shape n_blocks = block_size ** 2 if reverse: x = tf.reshape(x, (b, h, w, n_blocks, c // n_blocks)) else: x = tf.reshape(x, (b, h, w, c // n_blocks, n_blocks)) x = tf.transpose(x, (0, 1, 2, 4, 3)) x = tf.reshape(x, (b, h, w, c)) return x ``` | **Torch implementation** ```python class SpaceToDepth(nn.Module): def __init__(self, block_size): super().__init__() self.block_size = block_size def forward(self, input): x = F.pixel_unshuffle(input, self.block_size) x = channel_interleave2d(x, self.block_size, reverse=False) return x def channel_interleave2d(x: torch.Tensor, block_size: int, reverse: bool) -> torch.Tensor: b, c, h, w = x.shape n_blocks = block_size ** 2 if reverse: x = x.view(b, n_blocks, c // n_blocks, h, w) else: x = x.view(b, c // n_blocks, n_blocks, h, w) x = x.transpose(1, 2).reshape(b, c, h, w) return x ``` **Tensorflow converter** ```python @nobuco.converter(SpaceToDepth, channel_ordering_strategy=ChannelOrderingStrategy.FORCE_TENSORFLOW_ORDER) def converter_space_to_depth(self, input: torch.Tensor): return lambda input: tf.nn.space_to_depth(input, self.block_size) ``` |
Going dynamic
Control flows
Introducing python control flow statements into the compute graph is no easy feat.
Tensorflow can do so via tf.autograph, but at a cost of system's complexity and with some notable limitations.
Stuff like that is way above Nobuco's paygrade, so the following module cannot be properly handled without human intervention.
class ControlIf(nn.Module):
def __init__(self):
super().__init__()
self.conv_pre = nn.Conv2d(3, 16, kernel_size=(1, 1))
self.conv_true = nn.Conv2d(16, 32, kernel_size=(1, 1))
self.conv_false = nn.Conv2d(16, 32, kernel_size=(1, 1))
self.conv_shared = nn.Conv2d(32, 32, kernel_size=(1, 1))
def forward(self, x):
x = self.conv_pre(x)
if x.mean() > 0:
x = self.conv_true(x)
x = torch.tanh(x)
x = self.conv_shared(x)
x = x + 1
else:
x = self.conv_false(x)
x = torch.sigmoid(x)
x = self.conv_shared(x)
x = x - 1
x = self.conv_shared(x)
return xOf course, it's possible to translate the dynamic module into a Tensorflow layer
(don't forget to decorate it with @tf.function for autograph to kick in).
But what if it contains inner modules, do you replicate them in Tensorflow all by hand?
Not unless you want to!
Just convert them separately and use the resulting graphs inside the parent layer.
class ControlIfKeras(tf.keras.layers.Layer):
def __init__(self, conv_pre, conv_true, conv_false, conv_shared, *args, **kwargs):
super().__init__(*args, **kwargs)
self.conv_pre = conv_pre
self.conv_true = conv_true
self.conv_false = conv_false
self.conv_shared = conv_shared
def get_config(self):
config = super().get_config()
config.update({
"conv_pre": self.conv_pre,
"conv_true": self.conv_true,
"conv_false": self.conv_false,
"conv_shared": self.conv_shared,
})
return config
@tf.function
def call(self, x):
x = self.conv_pre(x)
if tf.reduce_mean(x) > 0:
x = self.conv_true(x)
x = tf.tanh(x)
x = self.conv_shared(x)
x = x + 1
else:
x = self.conv_false(x)
x = tf.sigmoid(x)
x = self.conv_shared(x)
x = x - 1
x = self.conv_shared(x)
return x
@nobuco.converter(ControlIf, channel_ordering_strategy=ChannelOrderingStrategy.FORCE_TENSORFLOW_ORDER)
def converter_ControlIf(self, x):
order = ChannelOrder.TENSORFLOW
kwargs = {'inputs_channel_order': order, 'outputs_channel_order': order, 'return_outputs_pt': True}
conv_pre, out_pre = nobuco.pytorch_to_keras(self.conv_pre, [x], **kwargs)
conv_true, out_true = nobuco.pytorch_to_keras(self.conv_true, [out_pre], **kwargs)
conv_false, out_false = nobuco.pytorch_to_keras(self.conv_false, [out_pre], **kwargs)
conv_shared, _ = nobuco.pytorch_to_keras(self.conv_shared, [out_true], **kwargs)
layer = ControlIfKeras(conv_pre, conv_true, conv_false, conv_shared)
return layer
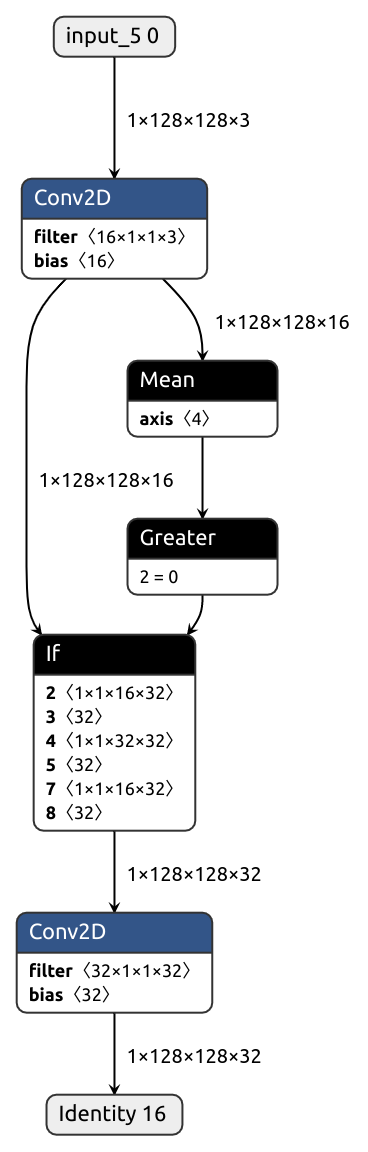
See examples for other ways to convert control flow ops.
Dynamic shapes
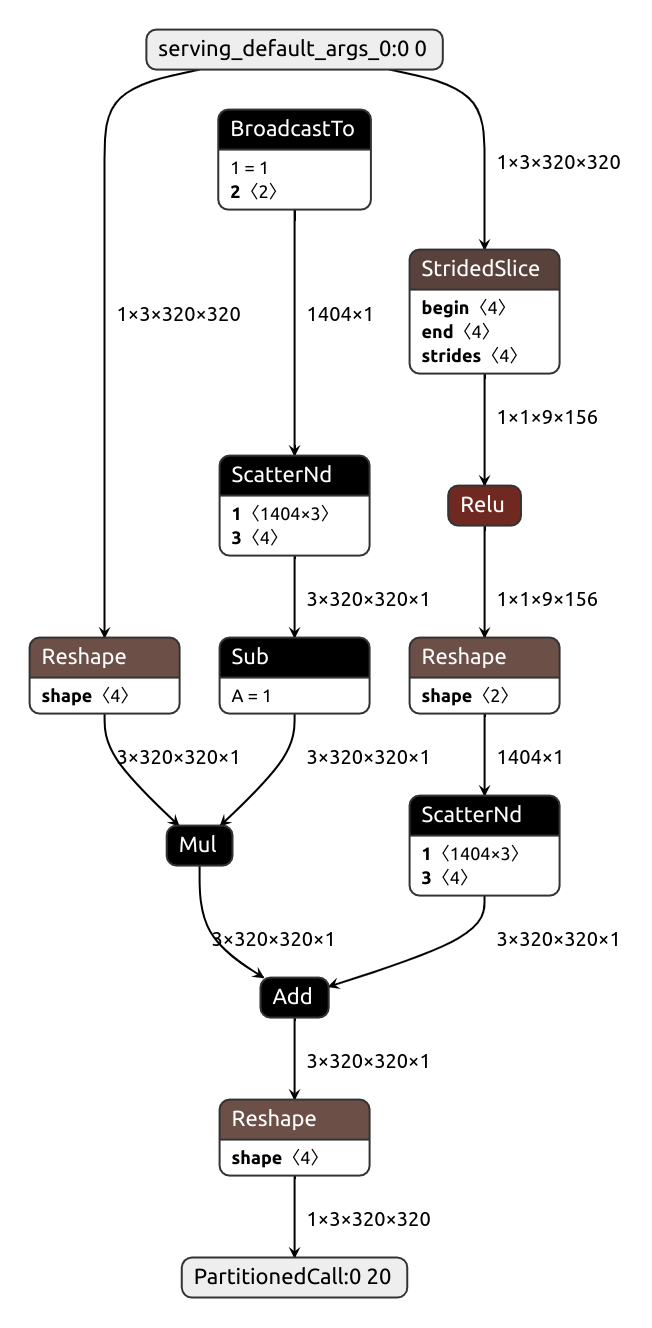
What if we wanted our module to accept images of arbitrary height and width? Can we have that? Let's try:
class DynamicShape(nn.Module):
def __init__(self):
super().__init__()
self.conv = nn.Conv2d(3, 16, kernel_size=(1, 1))
def forward(self, x):
x = self.conv(x)
# Produces static shape
b, c, h, w = x.shape
x = x[:, :, h//3:, w//3:]
return x
input = torch.normal(0, 1, size=(1, 3, 128, 128))
pytorch_module = DynamicShape().eval()
keras_model = nobuco.pytorch_to_keras(
pytorch_module,
args=[input],
input_shapes={input: (None, 3, None, None)}, # Annotate dynamic axes with None
inputs_channel_order=ChannelOrder.TENSORFLOW,
outputs_channel_order=ChannelOrder.TENSORFLOW,
)Something's not right. We don't see shape extraction ops in the debug output or the graph:
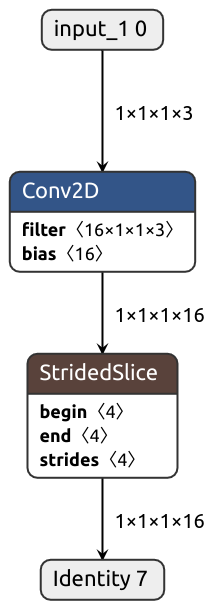
That's not surprising, actually.
In Pytorch, tensor shape is a tuple of regular integers, not tensors, so it's quite difficult to track them.
nobuco.shape solves this problem.
This function returns tensors, much like tf.shape does:
# Allows for dynamic shape
b, c, h, w = nobuco.shape(x)
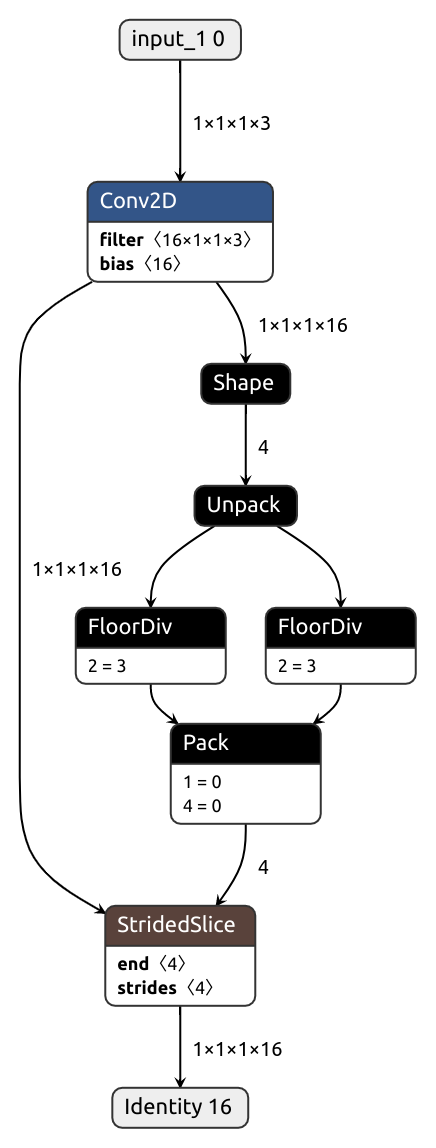
It's also possible to automatically substitute every .shape/.size call with nobuco.shape during the tracing phase by setting trace_shape flag:
keras_model = nobuco.pytorch_to_keras(
# ...
trace_shape=True
)No, that's too dynamic!
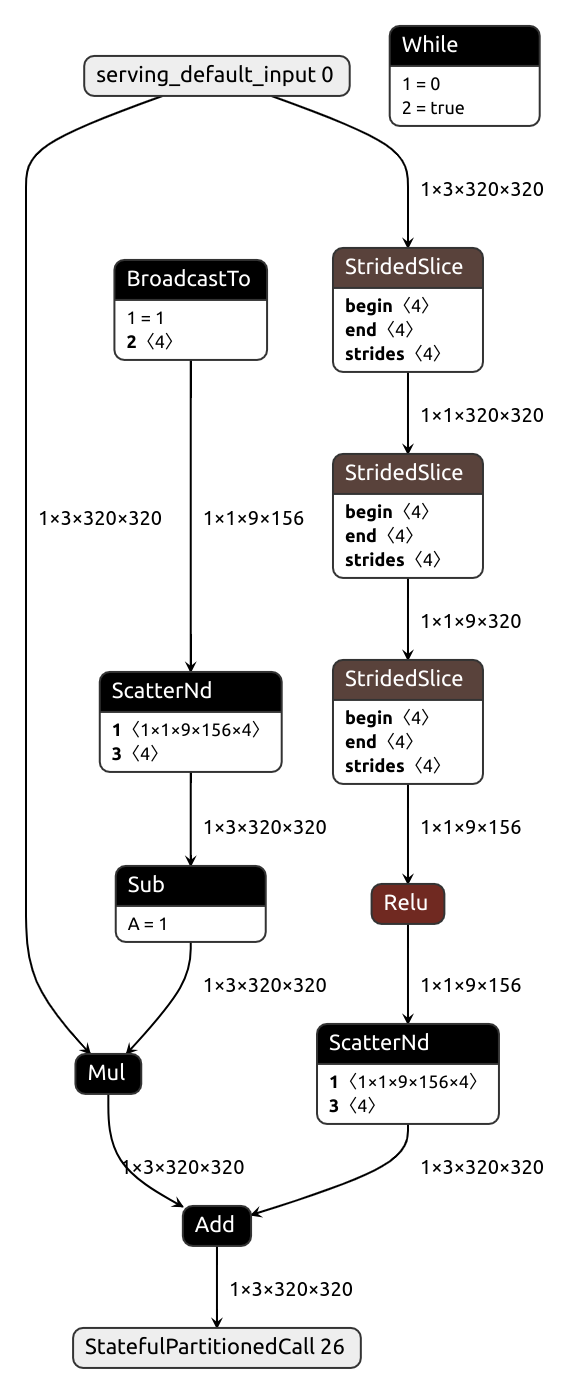
Forcing static crops
Sometimes, dynamic tensors appear against our will. They can also be very detrimental, especially when it comes to efficient inference. Let me show you.
Our Pytorch module here involves extracting a crop of a fixed size, something along these lines:
class CroppingModule(nn.Module):
def __init__(self, crop_height, crop_width):
super().__init__()
self.crop_height = crop_height
self.crop_width = crop_width
def forward(self, x, crop_x, crop_y):
_, _, h, w = x.shape
crop_x = (crop_x * (w - self.crop_width)).int()
crop_y = (crop_y * (h - self.crop_height)).int()
crop = x[:, :, crop_y: crop_y + self.crop_height, crop_x: crop_x + self.crop_width] # Poor way to crop
return crop
x = torch.normal(0, 1, size=(1, 3, 128, 128))
crop_y = torch.rand(())
crop_x = torch.rand(())
pytorch_module = CroppingModule(64, 32).eval()

INFO: Created TensorFlow Lite XNNPACK delegate for CPU.
WARNING: Attempting to use a delegate that only supports static-sized tensors with a graph that has dynamic-sized tensors (tensor#17 is a dynamic-sized tensor).
ERROR: Failed to apply XNNPACK delegate.TFLite fails to recognize the crop shape as static. Why? Consider this:
y0 = torch.tensor(())
y1 = y0 + h
crop = image[:, y0: y1]When we invoke image[:, y0: y1], the indexing operator ([]) has no way of knowing that y0 and y1 are related and y1 - y0 == const.
It wouldn't be a problem if instead of (y0, y1), the op accepted (y0, h) as cropping parameters, since h is a constant.
Is there such op? Meet torch.narrow:
crop = x.narrow(2, crop_y, self.crop_height).narrow(3, crop_x, self.crop_width) # Better way to crop

That's it, problem solved, the crop is statically-shaped now.
Alas, narrow only operates on one dimension at a time, so we have to apply it repeatedly.
tf.slice, however, has no such limitation. See where I'm going? It's custom converter time!
@nobuco.traceable
def get_crop(x, crop_y, crop_x, h, w):
return x[:, :, crop_y: crop_y + h, crop_x: crop_x + w]
@nobuco.converter(get_crop, channel_ordering_strategy=nobuco.ChannelOrderingStrategy.FORCE_TENSORFLOW_ORDER)
def converter_get_crop(x, crop_y, crop_x, h, w):
def func(x, crop_y, crop_x, h, w):
return tf.image.crop_to_bounding_box(x, crop_y, crop_x, h, w) # Calls tf.slice under the hood
return funccrop = get_crop(x, crop_y, crop_x, self.crop_height, self.crop_width) # Best way to crop

In-place operations
Nobuco can handle most situations where tensors are modified in-place. For instance, these will work just fine:
class MyModule(nn.Module):
def forward(self, x):
x[:, 1:2, 16:25, 8::2] *= 2
torch.relu_(x)
return xHowever, applying in-place operation to a slice yields incorrect result. What gives?
class MyModule(nn.Module):
def forward(self, x):
torch.relu_(x[:, 1:2, 16:25, 8::2])
return xYou see, Tensorflow graphs (and many other formats like ONNX) do not support in-place ops.
So when we take slice (x[:, 1:2, 16:25, 8::2]) in TF/ONNX, the result is not a view of the original tensor but a copy.
This copy is then passed to relu (which is not in-place either), and its result is not used anywhere.
As you can see above, the output tensors of __getitem__ and relu_ are grayed out, and these operations are excluded from the graph.
In fact, it's empty:
The easiest way of fixing this is to explicitly assign the result to the slice. Conveniently enough, most standard in-place operations in Pytorch do return their modified arguments as outputs.
class MyModule(nn.Module):
def forward(self, x):
x[:, 1:2, 16:25, 8::2] = torch.relu_(x[:, 1:2, 16:25, 8::2])
return xA little white lie: tracing mode
The flexibility and dynamic nature of Pytorch graphs can make it quite challenging to directly translate them not only to Tensorflow but ONNX as well. How are these types of problems solved for real-world models? Once again, we'll learn by example:
pytorch_module = torchvision.models.detection.ssdlite320_mobilenet_v3_large(weights=SSDLite320_MobileNet_V3_Large_Weights.DEFAULT).eval()
x = torch.rand(size=(1, 3, 320, 320))
keras_model = nobuco.pytorch_to_keras(
pytorch_module,
args=[x],
)Is that Nobuco's failure to handle in-place copy_? Yes, but there's more to the story.
Let's peek into the model's source code (set debug_traces=nobuco.TraceLevel.ALWAYS for easier navigation). Here's the culprit:
def batch_images(self, images: List[Tensor], size_divisible: int = 32) -> Tensor:
if torchvision._is_tracing():
# batch_images() does not export well to ONNX
# call _onnx_batch_images() instead
return self._onnx_batch_images(images, size_divisible) # <- Alternative ONNX-friendly implementation
max_size = self.max_by_axis([list(img.shape) for img in images])
stride = float(size_divisible)
max_size = list(max_size)
max_size[1] = int(math.ceil(float(max_size[1]) / stride) * stride)
max_size[2] = int(math.ceil(float(max_size[2]) / stride) * stride)
batch_shape = [len(images)] + max_size
batched_imgs = images[0].new_full(batch_shape, 0)
for i in range(batched_imgs.shape[0]):
img = images[i]
batched_imgs[i, : img.shape[0], : img.shape[1], : img.shape[2]].copy_(img) # <- In-place copy
return batched_imgsThis method is certainly not fit for tracing.
The model's authors knew it and provided an alternative implementation in case we'd want to export it to ONNX (works for Keras, too!).
torchvision._is_tracing() returns True whenever the model is being traced (e.g. invoked inside torch.onnx.export(...)).
This state is not directly controllable by the user, yet Nobuco can gaslight the model into thinking it's being traced by Pytorch itself:
keras_model = nobuco.pytorch_to_keras(
# ...
enable_torch_tracing=True
)Why is it not enabled by default, then?
You see, the other effect of tracing mode is equivalent to that of trace_shape=True: shape calls return tensors instead of ints.
Many Pytorch models were never meant to be converted to anything, and tracing mode may break them.
Nobuco tries to minimize silent/obscure errors and user's confusion, and not being too smart is a reasonable tradeoff.
Ad hoc modifications
Let's say, for illustrative purposes, that we prefer putting batchnorm before convolution.
We were sure TFLiteConverter would fuse these two linear operations into one.
Alas, it failed to meet our expectations.
Can we still get the fusion to work without re-training or messing around with the model checkpoint?
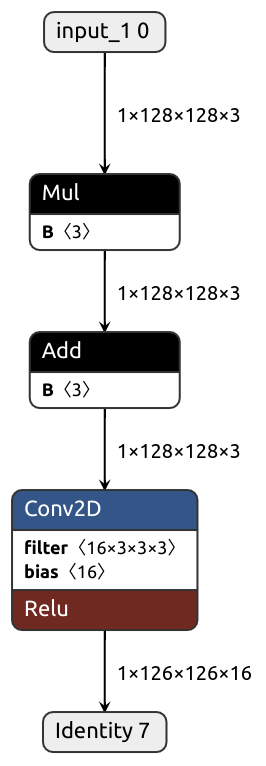
class FusibleModule(nn.Module):
def __init__(self):
super().__init__()
self.bn = nn.BatchNorm2d(3)
self.conv = nn.Conv2d(3, 16, kernel_size=(3, 3), padding=(0, 0))
self.act = nn.ReLU()
def forward(self, x):
x = self.bn(x)
x = self.conv(x)
x = self.act(x)
return x
Here's one way to do it:
- Wrap the two ops in a
Callable. Decorate it with@nobuco.traceable. - Make a custom converter for it which does the desired optimization.
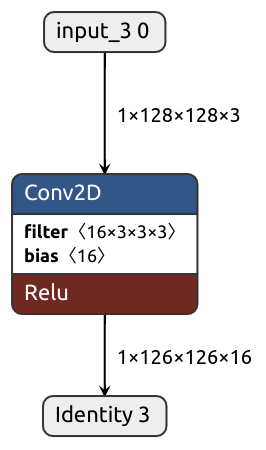
class FusibleModule(nn.Module):
# ...
@nobuco.traceable
def bn_conv(self, x):
x = self.bn(x)
x = self.conv(x)
return x
def forward(self, x):
x = self.bn_conv(x)
x = self.act(x)
return x@nobuco.converter(FusibleModule.bn_conv, channel_ordering_strategy=ChannelOrderingStrategy.FORCE_TENSORFLOW_ORDER)
def converter_bn_conv(self, x):
order = ChannelOrder.TENSORFLOW
bn, out_bn = nobuco.pytorch_to_keras(self.bn, [x], inputs_channel_order=order, outputs_channel_order=order, return_outputs_pt=True)
conv = nobuco.pytorch_to_keras(self.conv, [out_bn], inputs_channel_order=order, outputs_channel_order=order)
gamma, beta, moving_mean, moving_variance = bn.get_weights()
kernel, bias = conv.get_weights()
eps = self.bn.eps
'''
y = gamma * (x - moving_mean) / sqrt(moving_variance + eps) + beta
z = kernel * y + bias
=>
z = kernel_fused * x + bias_fused WHERE
kernel_fused = kernel * gamma / sqrt(moving_variance + eps)
bias_fused = -kernel_fused * moving_mean + kernel * beta + bias
'''
kernel_fused = kernel * (gamma / np.sqrt(moving_variance + eps))[None, None, :, None]
bias_fused = (-kernel_fused * moving_mean[None, None, :, None] + kernel * beta[None, None, :, None]).sum(axis=(0, 1, 2)).flatten() + bias
conv.set_weights([kernel_fused, bias_fused])
return lambda self, x: conv(x)
So we put a converter inside your converter
As we've learned, Nobuco gets confused when in-place operation is applied to a slice. There's a way to fix that, but let's not do it now. Instead, we'll use it as an excuse to explain the concept of nested converters. So, for this module, conversion will give us incorrect result:
class SliceReLU(nn.Module):
def forward(self, x):
# Gives incorrect result after conversion
torch.relu_(x[:, 1:2, 16:25, 8::2])
# That's the recommended approach, but we're not going for it now
# x[:, 1:2, 16:25, 8::2] = torch.relu_(x[:, 1:2, 16:25, 8::2])
return x
class MyModule(nn.Module):
def __init__(self):
super().__init__()
self.conv = nn.Conv2d(3, 3, kernel_size=(3, 3), padding=(1, 1))
def forward(self, x):
x = self.conv(x)
SliceReLU()(x)
return xWe've seen it's possible to invoke a Nobuco converter inside another Nobuco converter.
Can we embed some third-party converter? You bet! Why? Because it might just do what we need.
Let's consider the standard route: Pytorch -> ONNX -> Tensorflow, with the latter step done with onnx-tf.
This library likes transposing stuff so much, converting the whole graph with it may introduce intolerable inference overhead. Nonetheless, it does the job.
A sensible tradeoff would be to wrap the problematic operation into its own nn.Module and give it a special treat, while handling everything else with Nobuco.
import onnx
from onnx_tf.backend import prepare
@nobuco.converter(SliceReLU, channel_ordering_strategy=ChannelOrderingStrategy.FORCE_PYTORCH_ORDER, reusable=False)
def converter_SliceReLU(self, x):
model_path = 'slice_relu'
onnx_path = model_path + '.onnx'
# NB: onnx.export in implemented via tracing i.e. it may modify the inputs!
torch.onnx.export(self, (x,), onnx_path, opset_version=12, input_names=['input'],
dynamic_axes={'input': [0, 1, 2, 3]}
)
onnx_model = onnx.load(onnx_path)
tf_rep = prepare(onnx_model)
tf_rep.export_graph(model_path)
model = tf.keras.models.load_model(model_path)
return keras.layers.Lambda(lambda x: model(input=x))But was it worth it?
Let's cut to the chase, here's the numbers.
mobilenet_v3_large (26.8 Mb)
| nobuco | onnx_tf | speedup | |
|---|---|---|---|
| x86 (XNNPACK) | 11.1 ms | 14.7 ms | 1.3x |
| Arm CPU (XNNPACK) | 24.3 ms | 40.3 ms | 1.6x |
| Arm GPU (OpenCL) | 21.3 ms | 192.6 ms | 9x |
deeplabv3_resnet50 (158.5 Mb)
| nobuco | onnx_tf | speedup | |
|---|---|---|---|
| x86 (XNNPACK) | 1.25 s | 1.34 s | 1.07x |
| Arm CPU (XNNPACK) | 2.0 s | 2.7 s | 1.35x |
| Arm GPU (OpenCL) | 1.6 s | 2.6 s | 1.62x |
As we can see, redundant transpositions may completely ruin the performance, especially on a GPU. But that's not the only issue. Let's test this:
class SliceReLU(nn.Module):
def forward(self, x):
x[:, 1:2, 16:25, 8::2] = torch.relu_(x[:, 1:2, 16:25, 8::2])
return x| nobuco | onnx_tf | speedup | |
|---|---|---|---|
| x86 (XNNPACK) | 0.40 ms | 1.57 ms | 3.9x |
| Arm CPU | 4.6 ms | 2.9 ms | 0.6x |
| Arm CPU (XNNPACK) | 2.1 ms | FAIL | — |
| Arm GPU (OpenCL) | 21.8 ms | FAIL | — |
Again, the graph obtained with onnx_tf is much slower on x86 CPU.
Worse yet, on mobile processor, optimized TFLite delegates for both GPU and CPU failed.
No transpose ops were added this time, so who's to blame?
Just look what torch.onnx.export gives us:
 slice_relu.onnx
slice_relu.onnx
onnx_tf does a fair job optimizing the monstrosity of a graph it's given,
but combining consecutive slice ops seems too much to ask.
It also leaves out garbage nodes sometimes (note the free-floating While in this example).
Nobuco evades these types of problems by simply not dealing with onnx.
| slice_relu_nobuco | slice_relu_onnxtf |
|---|---|
|
|
|
Nobuco knowledge base
Don't want to convert anything but looking for a Tensorflow equivalent of a certain Pytorch node (operation or module)? Nobuco already implements quite a few node converters, most written in a concise and, hopefully, understandable way. These are located in nobuco/node_converters, and there's a utility function to help you find what you need:
node = torch.Tensor.repeat
# node = F.relu_
# node = nn.LSTM
location_link, source_code = nobuco.locate_converter(node)
print('Converter location:')
print(location_link)
print('Converter source code:')
print(source_code)Converter location:
File "/home/user/anaconda3/envs/nb/lib/python3.9/site-packages/nobuco/node_converters/tensor_manipulation.py", line 141
Converter source code:
@converter(torch.Tensor.repeat, channel_ordering_strategy=ChannelOrderingStrategy.MINIMUM_TRANSPOSITIONS)
def converter_repeat(self, *sizes):
def func(self, *sizes):
if get_channel_order(self) == ChannelOrder.TENSORFLOW:
sizes = permute_pytorch2keras(sizes)
return tf.tile(self, sizes)
return funcDeep dive
Aggressive transposition removal: fighting fire with fire
Despite trying its best, Nobuco may produce outrageously inefficient graphs:
```python
class MyModule(nn.Module):
def __init__(self):
super().__init__()
self.conv1 = nn.Conv1d(3, 3, kernel_size=1)
self.conv2 = nn.Conv1d(6, 6, kernel_size=1)
################################
# How it's translated to Keras #
################################
def forward(self, x): #
x = self.conv1(x) # <- Output is in TENSORFLOW order
#
x = x.reshape(-1, 6, 2) # <- Expects input in PYTORCH order, transposition needed
# Output is in PYTORCH order
#
x = self.conv2(x) # <- Expects input in TENSORFLOW order, transposition needed
return x #
```
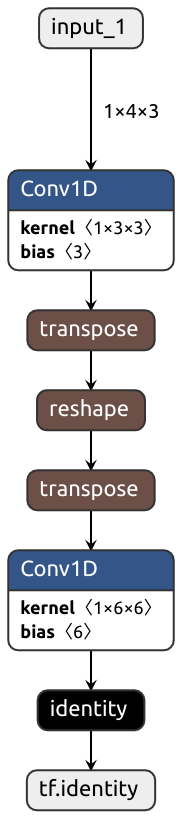
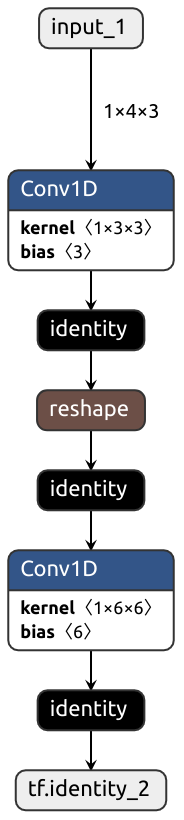
Acknowledgements
Slice assign converter is based on Zaccharie Ramzi's tf-slice-assign script.