ocean-ic
Create ocean initial conditions by regridding GODAS or ORAS4 reanalysis to MOM 0.25 degree, MOM 1 degree or NEMO grids.
Install
This tool is written in Python and depends a few different Python packages. It also depends on ESMF_RegridWeightGen program to perform regridding between non-rectilinear grids.
Download
Download ocean-ic:
$ git clone --recursive https://github.com/COSIMA/ocean-ic.git
$ cd ocean-ic
$ wget http://s3-ap-southeast-2.amazonaws.com/dp-drop/ocean-regrid/grid_defs.tar.gz
$ tar zxvf grid_defs.tar.gzOr see the section 'Tarballs' below.
Python dependencies
Use Anaconda as described below or an existing Python setup.
- Download and install Anaconda for your platform.
- Setup the Anaconda environment. This will download all the necessary Python packages.
$ cd ocean-ic $ conda env create -f regridder/regrid.yml $ source activate regrid
ESMF dependencies
Install ESMF_RegridWeightGen. ESMF releases can be found here.
There is a bash script regridder/contrib/build_esmf.sh which the testing system uses to build ESMF. This may be useful in addition to the ESMF installation docs.
Tarballs
Executable tarballs that include all Python dependencies but not ESMF_RegridWeightGen. It does not include the 'makeic_simple.py' described below, only more complete 'makeic.py'.
$ wget http://s3-ap-southeast-2.amazonaws.com/dp-drop/ocean-ic/release/makeic-0.0.4.tar.gz
$ tar zxvf makeic-0.0.4.tar.gz
$ export PATH=$(pwd)/makeic-0.0.4/:$PATH
$ makeic --helpUse
Download ORAS4 or GODAS reanalysis dataset, data can be found here:
- GODAS: http://www.esrl.noaa.gov/psd/data/gridded/data.godas.html
- ORAS4: ftp://ftp.icdc.zmaw.de/EASYInit/ORA-S4/monthly_orca1/
The examples below use preprepared inputs and outputs.
MOM 0.25 degree IC from GODAS
$ cd test
$ wget http://s3-ap-southeast-2.amazonaws.com/dp-drop/ocean-ic/test/test_data.tar.gz
$ tar zxvf test_data.tar.gz
$ cd test_data/input
$ export GRID_DEFS=../../../grid_defs
$ ../../../makeic.py GODAS $GRID_DEFS/pottmp.2016.nc $GRID_DEFS/pottmp.2016.nc \
pottmp.2016.nc salt.2016.nc \
MOM $GRID_DEFS/ocean_hgrid.nc $GRID_DEFS/ocean_vgrid.nc \
--model_mask $GRID_DEFS/ocean_mask.nc mom_godas_ic.nc
$ ncview mom_godas_ic.ncRather than 'makeic.py' there is also 'makeic_simple.py' which does not require the full paths to grid definitions are given.
$ cd test
$ wget http://s3-ap-southeast-2.amazonaws.com/dp-drop/ocean-ic/test/test_data.tar.gz
$ tar zxvf test_data.tar.gz
$ cd test_data/input
$ ../../../makeic_simple.py GODAS pottmp.2016.nc salt.2016.nc MOM mom_godas_ic.nc
$ ncview mom_godas_ic.ncTo verify your output:
$ mkdir -p test/example_output
$ cd test/example_output
$ wget http://s3-ap-southeast-2.amazonaws.com/dp-drop/ocean-ic/test/example_output/mom_godas_ic.nc
$ ncdiff mom_godas_ic.nc ../test_data/input/mom_godas_ic.nc diff.ncMOM 1 degree IC from GODAS
$ cd test
$ wget http://s3-ap-southeast-2.amazonaws.com/dp-drop/ocean-ic/test/test_data.tar.gz
$ tar zxvf test_data.tar.gz
$ cd test_data/input
$ export GRID_DEFS=../../../grid_defs
$ ../../../makeic.py GODAS $GRID_DEFS/pottmp.2016.nc $GRID_DEFS/pottmp.2016.nc \
pottmp.2016.nc salt.2016.nc \
MOM1 $GRID_DEFS/grid_spec.nc $GRID_DEFS/grid_spec.nc \
--model_mask $GRID_DEFS/grid_spec.nc mom1_godas_ic.nc
$ ncview mom_godas_ic.ncNote that the model name is now MOM1 instead of MOM as above. 'makeic_simple.py' can also be used as above.
NEMO IC from GODAS
Download the test data and set the GRID_DEFS environment variable as above.
$ cd test_data/input
$ ../../../makeic.py ORAS4 $GRID_DEFS/coordinates_grid_T.nc $GRID_DEFS/coordinates_grid_T.nc \
pottmp.2016.nc salt.2016.nc \
NEMO $GRID_DEFS/coordinates.nc $GRID_DEFS/data_1m_potential_temperature_nomask.nc \
nemo_godas_ic.ncMOM IC from ORAS4
Download the test data and set the GRID_DEFS environment variable as above.
$ cd test_data/input
$ ../../../makeic.py ORAS4 $GRID_DEFS/coordinates_grid_T.nc $GRID_DEFS/coordinates_grid_T.nc \
thetao_oras4_1m_2014_grid_T.nc so_oras4_1m_2014_grid_T.nc \
MOM $GRID_DEFS/ocean_hgrid.nc $GRID_DEFS/ocean_vgrid.nc \
--model_mask $GRID_DEFS/ocean_mask.nc mom_oras4_ic.ncNEMO IC from ORAS4
Download the test data as above.
$ cd test_data/input
$ ../../../makeic.py ORAS4 $GRID_DEFS/coordinates_grid_T.nc $GRID_DEFS/coordinates_grid_T.nc \
thetao_oras4_1m_2014_grid_T.nc so_oras4_1m_2014_grid_T.nc \
NEMO $GRID_DEFS/coordinates.nc $GRID_DEFS/data_1m_potential_temperature_nomask.nc \
nemo_oras4_ic.nc
$ ncview nemo_oras4_ic.ncAll of the above tests in one go
$ python -m pytest
$ ls test/test_data/output/How to use the output
MOM
Overwrite the output from the tool to the MOM initial condition file in the INPUT directory:
$ cp mom_oras4_ic.nc INPUT/ocean_temp_salt.res.ncNEMO
Overwrite the output from the tool to the NEMO initial condition file in the model run directory:
$ cp nemo_oras4_ic.nc data_1m_potential_temperature_nomask.nc
$ cp nemo_oras4_ic.nc data_1m_salinity_nomask.ncThen check the following namelist options:
&namrun ! parameters of the run
!-----------------------------------------------------------------------
ln_rstart = .false. ! start from rest (F) or from a restart file (T)
&namtsd ! data : Temperature & Salinity
!-----------------------------------------------------------------------
! ! file name ! frequency (hours) ! variable ! time interp. ! clim ! 'yearly'/ ! weights ! rotation ! land/sea mask !
! ! ! (if <0 months) ! name ! (logical) ! (T/F) ! 'monthly' ! filename ! pairing ! filename !
sn_tem = 'data_1m_potential_temperature_nomask', -1 ,'votemper' , .false. , .true. , 'yearly' , '' , '' , ''
sn_sal = 'data_1m_salinity_nomask' , -1 ,'vosaline' , .false. , .true. , 'yearly' , '' , '' , ''
ln_tsd_init = .true. ! Initialisation of ocean T & S with T &S input data (T) or not (F)
ln_tsd_tradmp = .false. ! damping of ocean T & S toward T &S input data (T) or not (F)
!-----------------------------------------------------------------------
&namtra_dmp ! tracer: T & S newtonian damping
!-----------------------------------------------------------------------
ln_tradmp = .false. ! add a damping termn (T) or not (F)Note that nudging / Newtownian damping (ln_tsd_tradmp and ln_tradmp) has been turned off and there is no time interpolation done on the input. The model output should then contain something like:
dta_tsd: deallocte T & S arrays as they are only use to initialize the runIf you do wish to do nudging / Newtownian damping then the initial condition must contain a time-series. One way to create this is using the ocean-nudge tool.
How it works
-
The reanalysis/obs dataset is regridded in the vertical to have the same depth and levels as the model grid. Linear interpolation is used for this. If the model is deeper than the obs then the deepest value is extended.
-
In the case of GODAS since the obs dataset is limited latitudinally it is extended to cover the whole globe. This is done based on nearest neighbours.
-
The obs dataset is then regridded onto the model grid using weights calculated with ESMF_RegridWeightGen. Various regridding schemes are supported includeing distance weighted nearest neighbour, bilinear and conservative.
-
The model land sea mask is applied and initial condition written out.
Limitations
- When using GODAS reanalysis the values at high latitudes are unphysical due to limited observations.
- Only 'cold-start' initial conditions are created consisting of temperature and salt fields. This means that the model will need to be spun up (possibly) with a reduced timestep. For example if you see output like the following from MOM it may be addressed by halving or quartering the timestep for a few months.
FATAL from PE 450: Error: temperature out of range with value 6.041772359369E+01 at (i,j,k) = ( 133, 520, 15), (lon,lat,dpt) = ( -246.8750, 5.3672, 164.1139 m)Example output
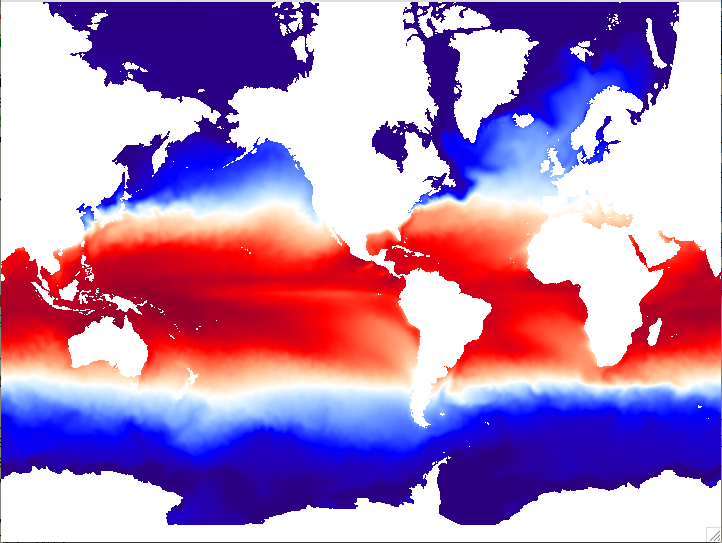
MOM IC temperature field based on ORAS4 reanalysis
MOM IC salt field based on GODAS reanalysis
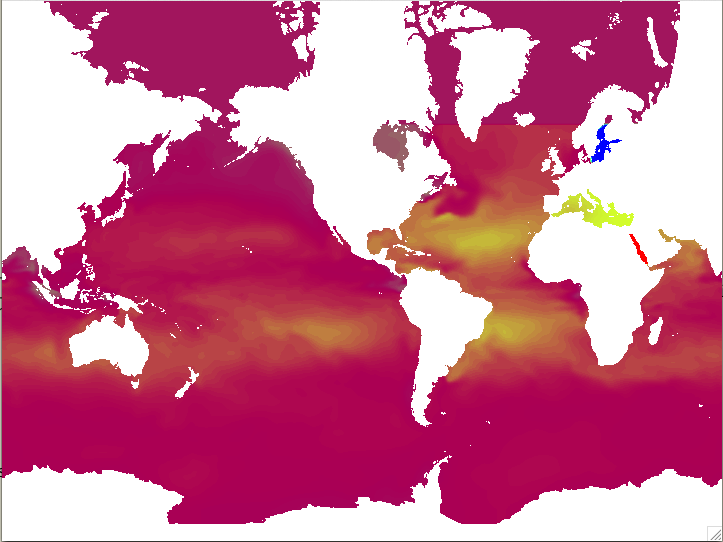
Note that because GODAS has a limited domain the salt in the Arctic has been filled with a 'representational value', in this case taken from the Bering Strait.
Developer notes
Package ocean-ic into a tarball using PyInstaller
Be aware of this issue https://github.com/pyinstaller/pyinstaller/issues/1781. It may be necessary to downgrade setuptools with the following command:
$ conda install setuptools==19.2First install pyinstaller:
$ pip install pyinstallerCreate release:
$ cd release
$ pyinstaller makeic.spec
$ mv dist/makeic ./makeic-x.x.x
$ tar czvf makeic-x.x.x.tar.gz makeic-x.x.xUpload tarball to s3:
$ s3put -b dp-drop -p /short/v45/nah599/more_home/ ./makeic-x.x.x.tar.gz
$ s3cmd setacl --acl-public --guess-mime-type s3://dp-drop/ocean-ic/release/makeic-x.x.x.tar.gzDownload ocean-ic tarball and test
$ wget http://s3-ap-southeast-2.amazonaws.com/dp-drop/ocean-ic/release/makeic-0.0.4.tar.gz
$ tar zxvf makeic-0.0.4.tar.gz
$ export PATH=$(pwd)/makeic-0.0.4/:$PATH
$ makeic_simple --help
$ mkdir -p test
$ cd test/
$ wget http://s3-ap-southeast-2.amazonaws.com/dp-drop/ocean-ic/test/test_data.tar.gz
$ tar zxvf test_data.tar.gz
$ cd test_data/input
$ makeic_simple GODAS pottmp.2016.nc salt.2016.nc NEMO nemo_godas_ic.ncCompare to known output:
$ mkdir example_output
$ cd example_output
$ wget http://s3-ap-southeast-2.amazonaws.com/dp-drop/ocean-ic/test/example_output/nemo_godas_ic.nc
$ ncdiff nemo_godas_ic.nc ../nemo_godas_ic.nc diff.nc
$ ncview diff.nc