PACO "Partitioning Cost Optimization"
Source Release 0.10 Alpha December, 12, 2016
PACO is a tool to build solvers and optimize quality functions on graphs for community detection. PACO comes as both a C++ library and a support for Matlab/Octave mex function as well as a Python library.
Publications based on the use of PACO should cite:
"Modular structure of brain functional networks: breaking the resolution limit by Surprise" C. Nicolini and A. Bifone, Scientific Reports doi:10.1038/srep19250
"Community detection in weighted brain connectivity networks beyond the resolution limit." Nicolini C., Bordier C., Bifone A. Neuroimage. 2017 Feb 1;146:28-39. doi: 10.1016/j.neuroimage.2016.11.026 *
Requirements
In order to compile PACO you need: COMPULSORY:
- The
Clibraries ofigraphat least0.7.1 - A C/C++ compiler (tested on
g++ <=5.5andclang <= 800-0.38), not tested under any Windows compiler. - CMake to generate the Makefiles or projects.
OPTIONAL
- MATLAB with mex compiler
- Octave with all headers files installed (package
octave-devin Ubuntu) - Python with Cython extensions (tested on Cython >=0.20)
Compilation of simple command line paco_optimizer
In order to compile the paco_optimizer command line executable you must do:
$> git clone --recurse-submodules https://github.com/carlonicolini/paco
$> cd paco
$> mkdir build
$> cd build
$> cmake ..
$> makeand you will start the compilation. Otherwise if you have the tar.gz with the last release:
$> tar -zxvf paco_0_10_alpha.tar.gz
$> cd paco_0_10_alpha
$> mkdir build
$> cd build
$> cmake ..
$> makePACO supports some options for the compile-time that enable the generation of Matlab and Octave wrappers.
To enable them, specify the option -DMATLAB_SUPPORT or -DOCTAVE_SUPPORT when you run cmake:
Compilation of MATLAB mex file
In order to compile the paco_mx mex function you must do:
$> tar -zxvf paco_0_10_alpha.tar.gz
$> cd paco_0_10_alpha
$> mkdir build
$> cd build
$> cmake -DMATLAB_SUPPORT=True ..
$> makeIf you have newer versions of MATLAB (>R2017a) or non-standard installations of MATLAB it may be necessary to specify the MATLAB_ROOT variable during the creation of the Makefile.
If your installation of Matlab is /usr/local/MATLAB/R2017b/bin/matlab then you need to specify the MATLAB_ROOT as follows
$> cmake -DMATLAB_SUPPORT=True -DMATLAB_ROOT=/usr/local/MATLAB/R2017b ..
$> makeYou can check if Matlab is found in the output of CMake. For example this is an healthy output:
-- MATLAB include directory: /usr/local/MATLAB/R2016a/extern/include
-- Matlab include directories: /usr/local/MATLAB/R2016a/extern/include
-- Found components for IGRAPH
-- IGRAPH_INCLUDES = /usr/local/include/igraph
-- IGRAPH_LIBRARIES = /usr/local/lib/libigraph.so
-- IGRAPH_VERSION_STRING_LINE = #define IGRAPH_VERSION "0.7.1"
-- IGRAPH_VERSION_MAJOR_GUESS = 0
-- IGRAPH_VERSION_MINOR_GUESS = 7
-- IGRAPH_VERSION_PATCH_GUESS = 1
-- MEX Files extension ".mexa64"
-- ======== /usr/local/MATLAB/R2016a/bin/glnxa64/libmx.so;/usr/local/MATLAB/R2016a/bin/glnxa64/libmex.so;/usr/local/MATLAB/R2016a/bin/glnxa64/libmat.so;/usr/local/MATLAB/R2016a/bin/glnxa64/libut.so ======
-- Configuring done
-- Generating done
-- Build files have been written to: /home/user/paco/buildCompilation of OCTAVE mex file
In order to compile the paco_oct mex function you must do:
$> tar -zxvf paco_0_10_alpha.tar.gz
$> cd paco_0_10_alpha
$> mkdir build
$> cd build
$> cmake -DOCTAVE_SUPPORT=True ..
$> makeCompilation of PYTHON module
In order to compile the pypaco python module you must do:
$> tar -zxvf paco_0_10_alpha.tar.gz
$> cd paco_0_10_alpha
$> mkdir build
$> cd build
$> cmake -DPYTHON_SUPPORT=True ..
$> makeSpecify the igraph libraries
PACO depends heavily on igraph-0.7.1. If you didn't install the igraph 0.7.1 libraries in the standard location (for example you don't have administrative privileges for your computer), you need to specify them at Cmake time with the options
-DCMAKE_EXTRA_LIBRARIES="PATH_TO_YOUR_IGRAPH_DIRECTORY_WHERE_LIBS_ARE_STORED"
-DCMAKE_EXTRA_INCLUDE="PATH_TO_YOUR_IGRAPH_DIRECTORY_WHERE_INCLUDE_ARE_STORED"For example the files libigraph.a libigraph.la libigraph.so libigraph.so.0 libigraph.so.0.0.0 are stored in $HOME/lib and the headers files igraph***.h are stored in $HOME/include/igraph. In this cased you need to specify CMake where these two folders are in this way:
$> cmake -DCMAKE_EXTRA_LIBRARIES=$HOME/lib/ -DCMAKE_EXTRA_INCLUDES=$HOME/include ..and then run make as usual.
Usage of PACO
Usage of command line optimizer
Once compiled, you can see the instructions of paco_optimizer
$> ./paco_optimizer
Usage: paco_optimizer graph_file [options]
graph_file the file containing the graph. Accepted formats are pajek, graph_ml, adjacency matrix or
edges list (the ncol format), additionally with a third column with edge weights
Options:
-q [quality]
0 Binary Surprise
1 Significance
2 Asymptotic Surprise
3 Infomap
-m [method]: 0 Agglomerative Optimizer
1 Random
2 Simulated Annealing
-V [report_level] ERROR=0, WARNING=1, INFO=2, DEBUG=3, DEBUG1=4, DEBUG2=5, DEBUG3=6, DEBUG4=7
-S [seed] specify the random seed, default time(0)
-b [bool] wheter to start with initial random cluster or every node in its community
-r [repetitions], number of repetitions of PACO, default=1
-p [print solution]The supported graph file formats are PAJEK (.net extension), GRAPHML (.gml extension), full adjacency matrix (text file with the values of adjacency matrix with .adj extension), or edges list (text file with edges list as two column file for unweighted or three column file for weighted edges, extension .ncol)
Usage of PACO mex file under MATLAB:
Once compiled, check that the matlab mex file paco_mx.* exists in your PACO folder. Start a MATLAB session and issue:
> addpath('folder_of_paco_mex_file')
> paco_mxThis command with no arguments will show you all the PACO options and an example usage.
>> paco_mx
PACO PArtitioning Cost Optimization.
[membership, qual] = paco(W);
Input:
W: an undirected weighted network with positive edge weights. Negative edge weights are not seen as edges and therefore discarded. Remember to use real matrices, logical matrices throw error.
Output:
membership: the membership vector that represents the community which every vertex belongs to after quality optimization.
qual: the current quality of the partition.
Options:
paco accepts additional arguments to control the optimization process
[m, qual] = paco(W,'method',val);
val is one of the following integers: {0,1,2}:
0: Agglomerative
1: Random
2: Annealing (EXPERIMENTAL)
[m, qual] = paco(W,'quality',val);
val is one of the following integers: {0,1,2,3}:
0: Surprise (discrete)
1: Significance
2: AsymptoticSurprise
3: Infomap
[m, qual] = paco(W,'nrep',val)
val is the number of repetitions to run over which to choose the best quality value (the lowest for Infomap, the highest for the other methods
[m, qual] = paco(W,'seed',val)
val is a specific random seed to the algorithm, in order to have reproducible results.
Example:
>> A=rand(100,100); A=(A+A')/2; A=A.*(A>0.5);
% Run Asymptotical Surprise optimization on A for 1000 repetitions and return the highest Surprise
partition membership together with the quality value
>> [memb, qual] = paco(A,'method',2,'nrep',1000);
Error using paco_mx
Error at argument: 0: Not enough input arguments.Usage of PACO mex file under OCTAVE:
Once compiled, check that the matlab mex file paco_mx.* exists in your PACO folder. Start an OCTAVE session and issue:
> addpath('folder_of_paco_mex_file')
> paco_octThis command with no arguments will show you all the PACO options and an example usage under OCTAVE. The syntax under MATLAB and OCTAVE is the same, only the filenames are different to avoid MATLAB/OCTAVE interpreter confusion.
P.S. Always remember not to compile MATLAB and OCTAVE mex files together, as the mex.h header can be misunderstood from the build system and strange problems may appear.
Usage of PACO as a Python module
Once compiled import PACO as a Python module:
>> from pypaco import pacoPACO Usage
Usage:
[membership, quality] = paco(A, **kwargs)
Args:
graph_rep: Adjacency matrix of the graph as a numpy 2D array, symmetric, binary or weighted. Otherwise an edgelist with m rows and 2 or 3 columns representing nodes indices
Kwargs:
quality: quality function to maximize
0: Surprise
1: Significance
2: Asymptotic Surprise
3: Infomap
opt_method: Optimization method
0: Agglomerative,
1: Random,
2: SimulatedAnnealing,
3: Infomap
nreps: number of repetitions of PACO (can increase the quality of the partition), (default 1).
seed: random seed for randomization (integer value)
save_solution: 1 to save ongoing solution, 0 otherwise (default 0)
Out:
membership: a list of vertices community membership
quality: the partition quality valueExample: passing graph as adjacency matrix
import numpy as np
from pypaco import paco
G = nx.karate_club_graph()
A = nx.to_numpy_matrix(G)
[membership,quality] = paco(A, quality=0, nreps=10)Example: passing graph as edges list
import numpy as np
from pypaco import paco
G = nx.karate_club_graph()
E = np.array(G.edges()).astype(float64) # E must be a numpy array of doubles, not a list
[membership,quality] = paco(E, quality=0, nreps=10)Example: passing graph as weighted edges list
import numpy as np
from pypaco import paco
G = nx.karate_club_graph()
E = np.array(G.edges())
# Generate weights on the edges, in values between 0 and 10-1
W = np.random.randint(10,size=[G.number_of_edges(),1])
EW = np.concatenate((E,W),axis=1).astype(float)
[membership,quality] = paco(EW, quality=2, nreps=10)Obtaining igraph in Ubuntu 12.04 or greater
We suggest compiling igraph from source, since PACO needs igraph>=0.7.1.
Go to www.igraph.org/c and follow the documentation for the compilation using the autogen script they provide, otherwise follow the following instructions:
Compilation and installation of igraph on Ubuntu 12.04 or greater
- Download igraph-0.7.1 source code:
$> cd
$> wget http://igraph.org/nightly/get/c/igraph-0.7.1.tar.gz- Extract the code under your home folder:
$> tar -zxvf igraph-0.7.1.tar.gz- Install the necessary libs, under Ubuntu:
$> sudo apt-get install build-essential libxml2-dev zlib1g-dev- Try to configure and compile igraph:
$> cd igraph-0.7.1/
$> ./configure
$> make -
If you have many processors on your computer, make in parallel mode, instead of
makejust issuemake -j8for example to compile with 8 cores. -
Check if installation went fine:
$> make check- If tests are passed, then install it. I usually install igraph under
/usr/lib/. To do that I callmake installwith administrator privileges.
$> sudo make install- If you don't have administrator privileges you can compile igraph under your home folder.
In the rest of the notes we assume that the Igraph include directory is /usr/local/include/igraph.
Obtaining igraph in OSX
We strongly suggest to obtain igraph in OSX using Homebrew.
$> ruby -e "$(curl -fsSL https://raw.githubusercontent.com/Homebrew/install/master/install)"
$> brew tap homebrew/science
$> brew install igraphThe CMAKE will then find the installed dependencies correctly.
Windows support:
The support in Windows is experimental. I've managed to compile it smoothly with the following settings:
- Windows 7
- CMake 3.1
- Visual Studio 2013 (64 bit)
If you need to enable the MATLAB_SUPPORT you may need a recent version of Matlab. I'm using
- MATLAB R2016a
this version only supports 64 bit systems, so the mex file provided is 64 bits.
Instructions for compilation on Windows
You first need to compile the Visual Studio project
-
Compile igraph-0.7.1 for Visual Studio with x64 architecture:
- Download it from http://igraph.org/nightly/get/msvc/igraph-0.7.1-msvc.zip and extract it in the root PACO folder. Open the solution and compile it in Release mode with x64 architecture.
-
Run CMake in the PACO folder and set Visual Studio 2013 x64 toolset. Press configure, then tick the option MATLAB_SUPPORT and check that the Matlab libraries are correctly found, like in this picture
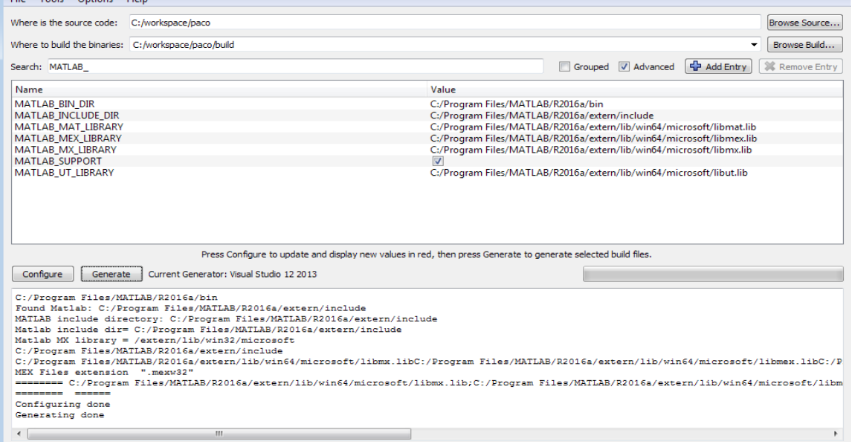
- Once the solution is generated, go to Visual Studio and compile the project
paco_mx. The mex file should go underpaco/build/Release/x64/folder.
FAQ
I can compile PACO but Matlab crashes with linker problems
It may happen on the latest versions of Matlab (>R2017a) that you get the following error:
Invalid MEX-file '/home/user/paco_mx.mexa64':
Missing symbol '_ZNKSt7__cxx1112basic_stringIcSt11char_traitsIcESaIcEE5rfindEcm' required by
'/usr/lib/x86_64-linux-gnu/libigraph.so.0->/home/user/libPACO.so->/home/user/paco_mx.mexa64'
Missing symbol '_ZNKSt7__cxx1112basic_stringIcSt11char_traitsIcESaIcEE7compareEPKc' required by
'/usr/lib/x86_64-linux-gnu/libigraph.so.0->/home/user/libPACO.so->/home/user/paco_mx.mexa64'
Missing symbol '_ZNSt7__cxx1112basic_stringIcSt11char_traitsIcESaIcEE10_M_replaceEmmPKcm' required by
'/usr/lib/x86_64-linux-gnu/libigraph.so.0->/home/user/libPACO.so->/home/user/paco_mx.mexa64'
Missing symbol '_ZNSt7__cxx1112basic_stringIcSt11char_traitsIcESaIcEE14_M_replace_auxEmmmc' required by
'/usr/lib/x86_64-linux-gnu/libigraph.so.0->/home/user/libPACO.so->/home/user/paco_mx.mexa64'
Missing symbol '_ZNSt7__cxx1112basic_stringIcSt11char_traitsIcESaIcEE9_M_appendEPKcm' required by
'/usr/lib/x86_64-linux-gnu/libigraph.so.0->/home/user/libPACO.so->/home/user/paco_mx.mexa64'
Missing symbol '_ZNSt7__cxx1112basic_stringIcSt11char_traitsIcESaIcEE9_M_assignERKS4_' required by
'/usr/lib/x86_64-linux-gnu/libigraph.so.0->/home/user/libPACO.so->/home/user/paco_mx.mexa64'
Missing symbol '_ZNSt7__cxx1112basic_stringIcSt11char_traitsIcESaIcEE9_M_createERmm' required by
'/usr/lib/x86_64-linux-gnu/libigraph.so.0->/home/user/libPACO.so->/home/user/paco_mx.mexa64'To fix this error you need to install matlab-support and rename the gcc libraries when asked:
sudo apt-get install matlab-support Linking libstdc++.so.6 problem
-
Matlab is not finding the correct gcc libraries on Linux. The solution here is to install the package matlab-support
sudo apt-get install matlab-support
when asked to rename the GCC libraries, press YES and continue. This will fix the uncorrect symbolic links. If this approach doesn't work for you, continue to the next step.
- I can compile PACO for MATLAB but after calling
paco_mx, MATLAB prompts me with the following error message:
Invalid MEX-file '~/paco_mx.mexa64': /usr/local/MATLAB/R2015a/bin/glnxa64/../../sys/os/glnxa64/libstdc++.so.6: version `GLIBCXX_3.4.21' not found (required by ...This problem means that the libstdc++.so.6 inside the Matlab library folder is pointing to a version of libstdc++ older than the system one, usually stored in /usr/lib/x86_64 folder.
To solve the issue you need to redirect the symbolic links in the MATLAB folder to the systemwise libstdc++. Hereafter we assume the MATLAB folder to be /usr/local/MATLAB/R2015a and the system to be some Linux variant.
Two of the symlinks for libraries need to be changed:
$> cd /usr/local/MATLAB/R2015a/sys/os/glnxa64
$> ls -lThe sym links for libstdc++.so.6 and libgfortran.so.3 should point to versions in /usr/lib, not local ones.
Before changing this libraries, first make sure g++-4.4 and libgfortran3are installed :
$> sudo apt-get install g++-4.4 libgfortran3Now, modify the symlinks:
$> sudo ln -fs /usr/lib/x86_64-linux-gnu/libgfortran.so.3.0.0 libgfortran.so.3
$> sudo ln -fs /usr/lib/gcc/x86_64-linux-gnu/4.4/libstdc++.so libstdc++.so.6This command makes the libstdc++.so.6 point to the g++-4.4 libstdc++ library.
For other informations take a look at http://dovgalecs.com/blog/matlab-glibcxx_3-4-11-not-found/ or https://github.com/RobotLocomotion/drake/issues/960 which are very similar problems.