FirstBreaksPicking
This project is devoted to pick waves that are the first to be detected on a seismogram (first breaks, first arrivals). Traditionally, this procedure is performed manually. When processing field data, the number of picks reaches hundreds of thousands. Existing analytical methods allow you to automate picking only on high-quality data with a high signal / noise ratio.
As a more robust algorithm, it is proposed to use a neural network to pick the first breaks. Since the data on adjacent seismic traces have similarities in the features of the wave field, we pick first breaks on 2D seismic gather, not individual traces.
Examples
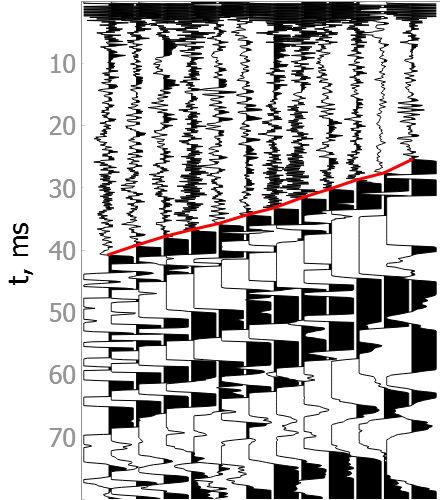
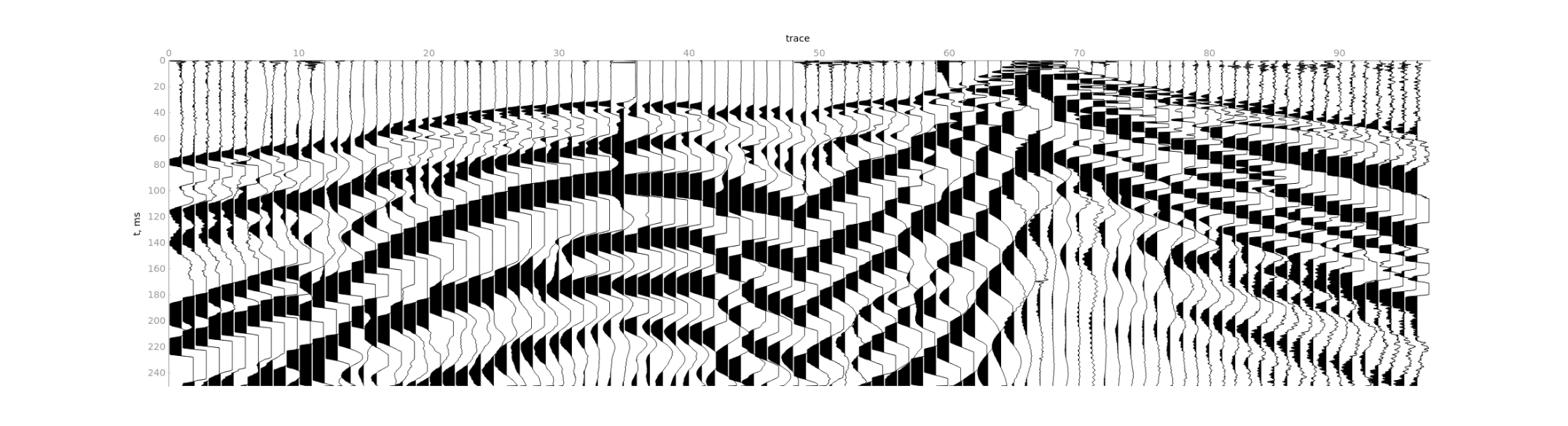
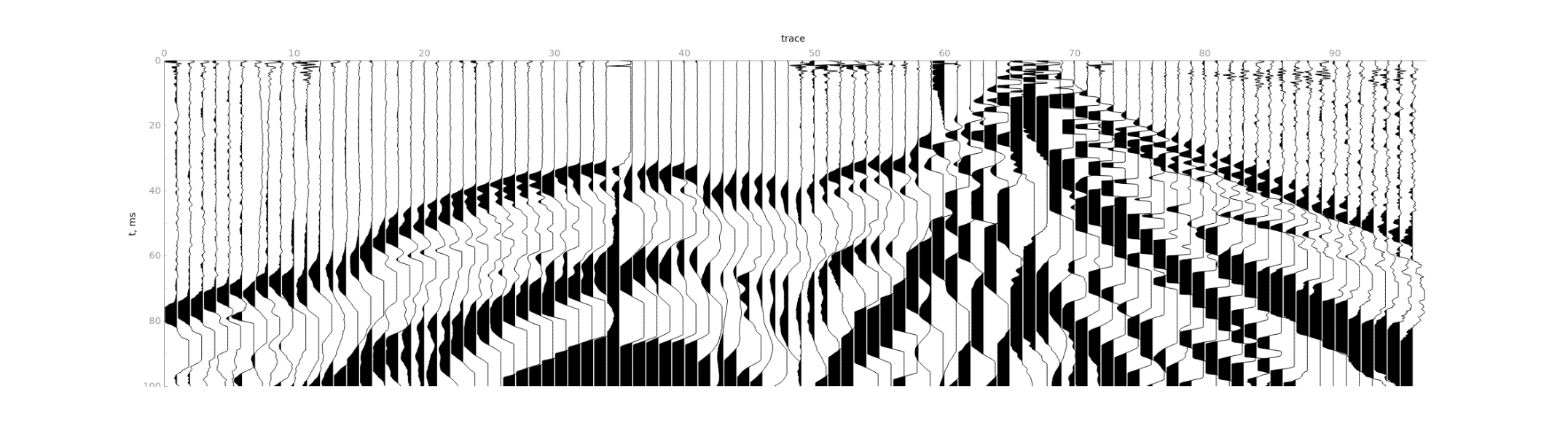
In the drop-down menus below you will find examples of our model picking on good quality data as well as noisy ones.
It is difficult to validate the quality of the model on noisy data, but they show the robustness of the model to various types of noise.
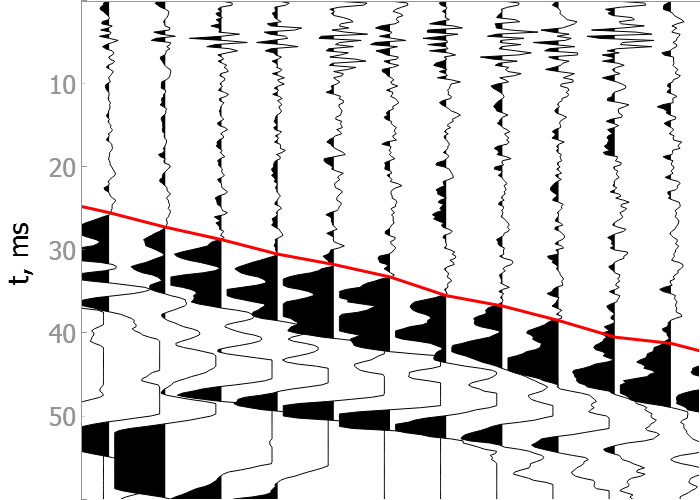
Good quality data
  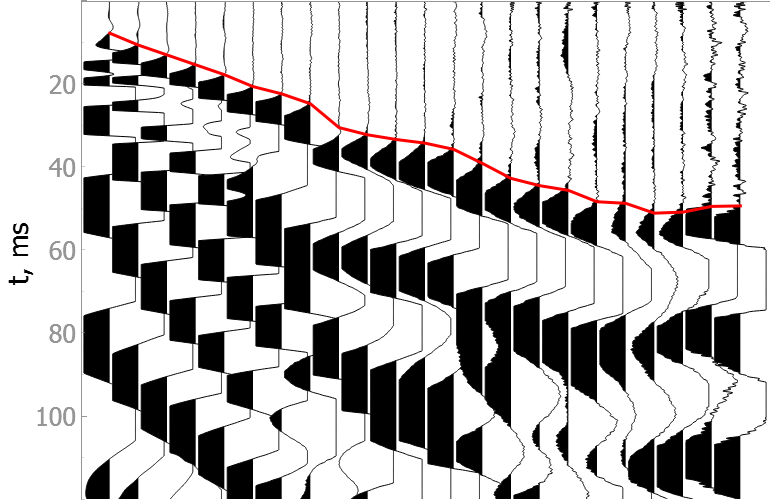 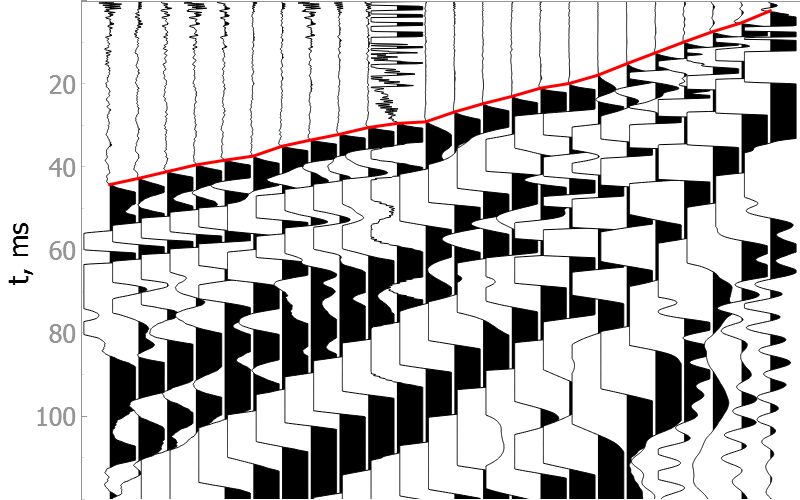   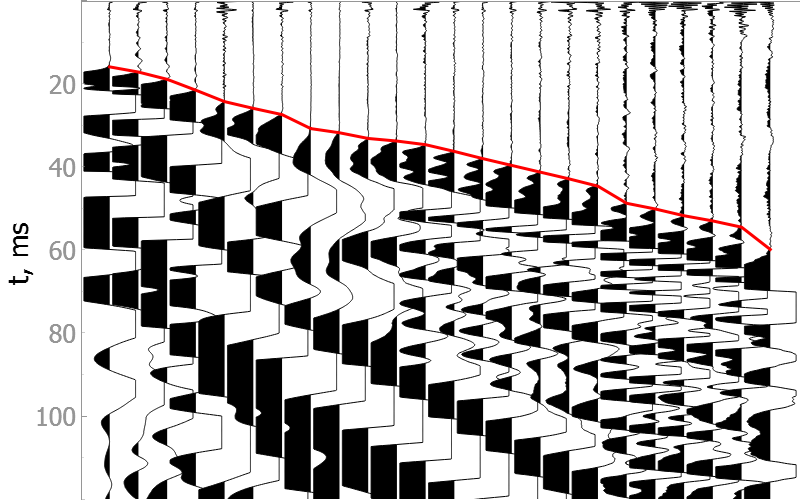 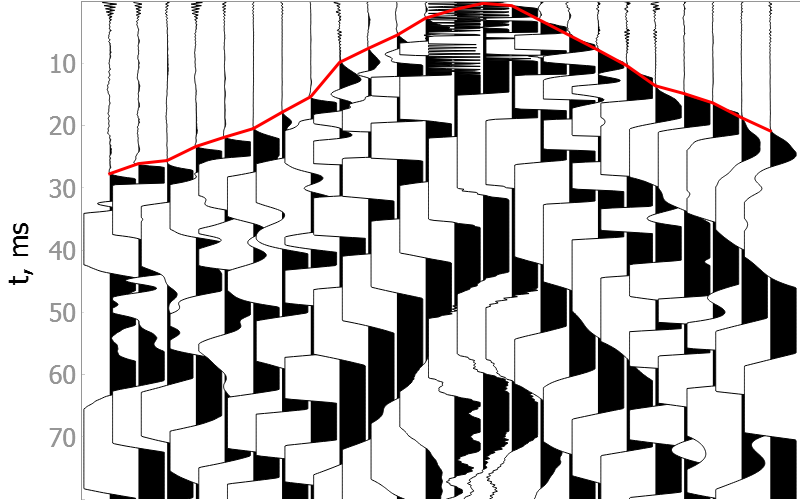  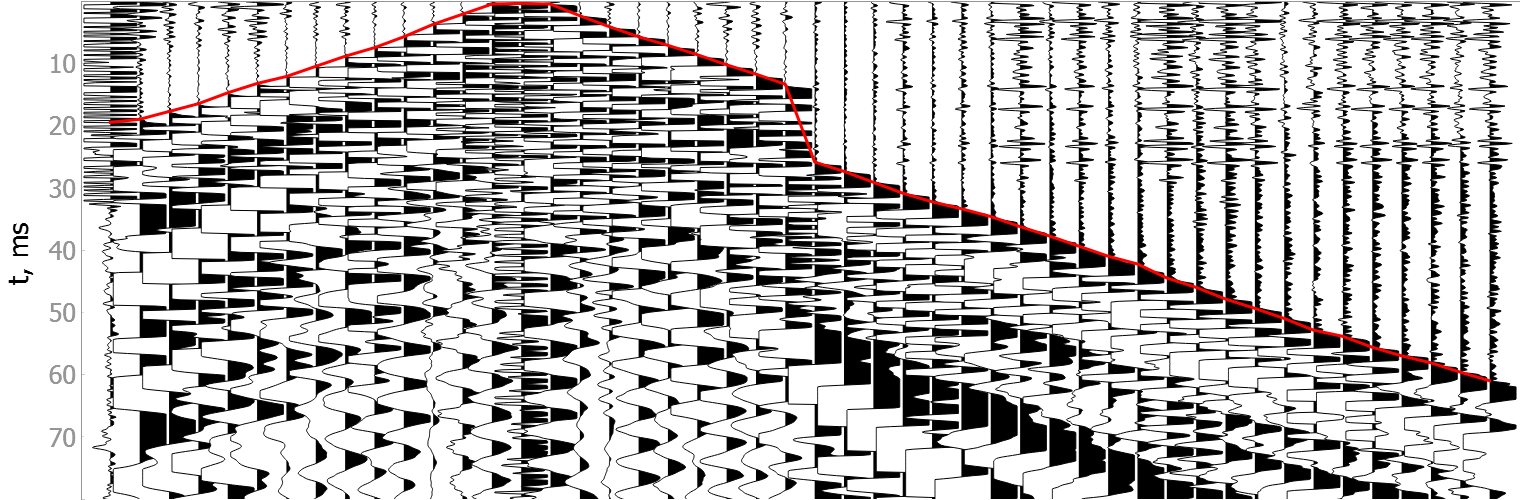 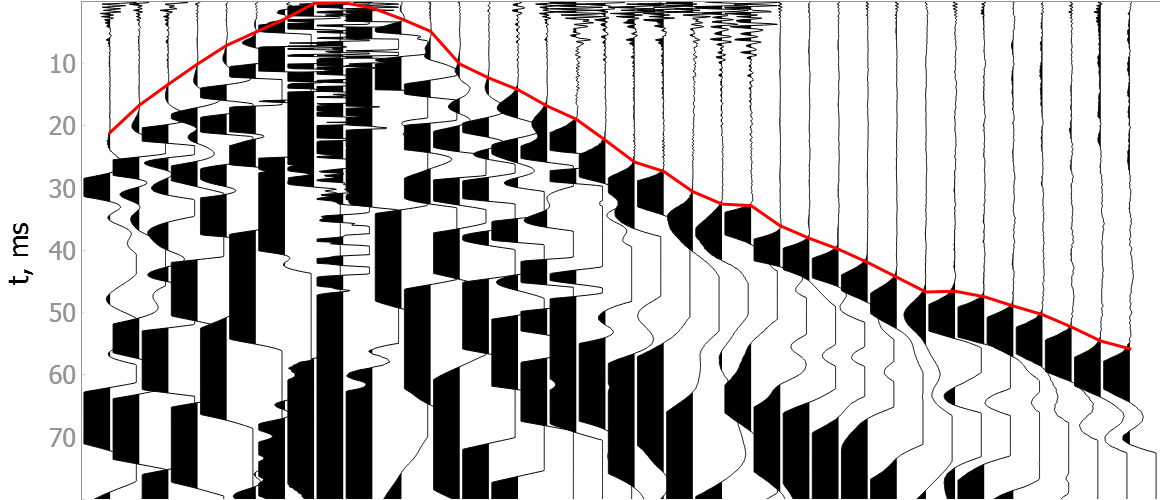  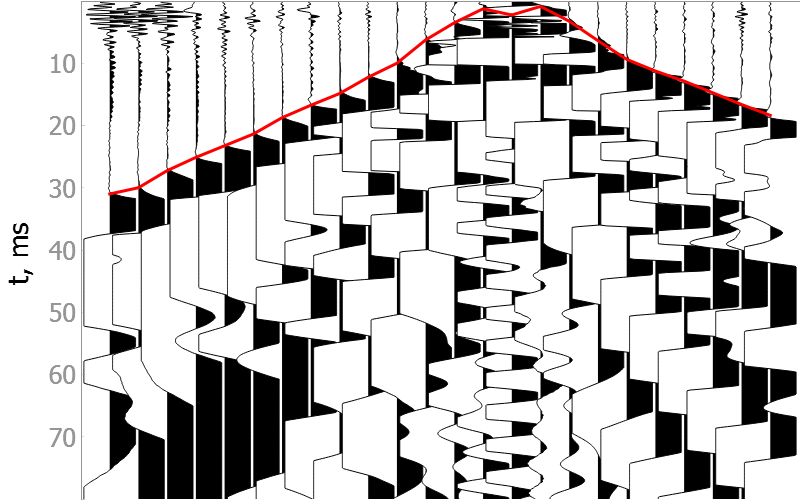 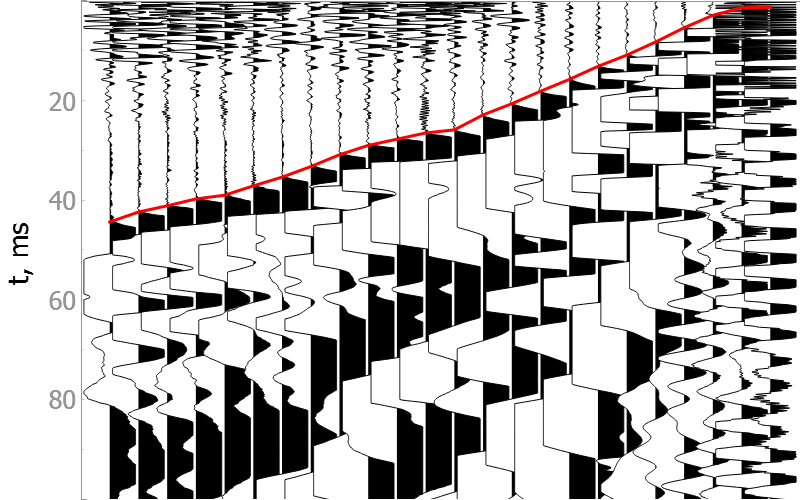 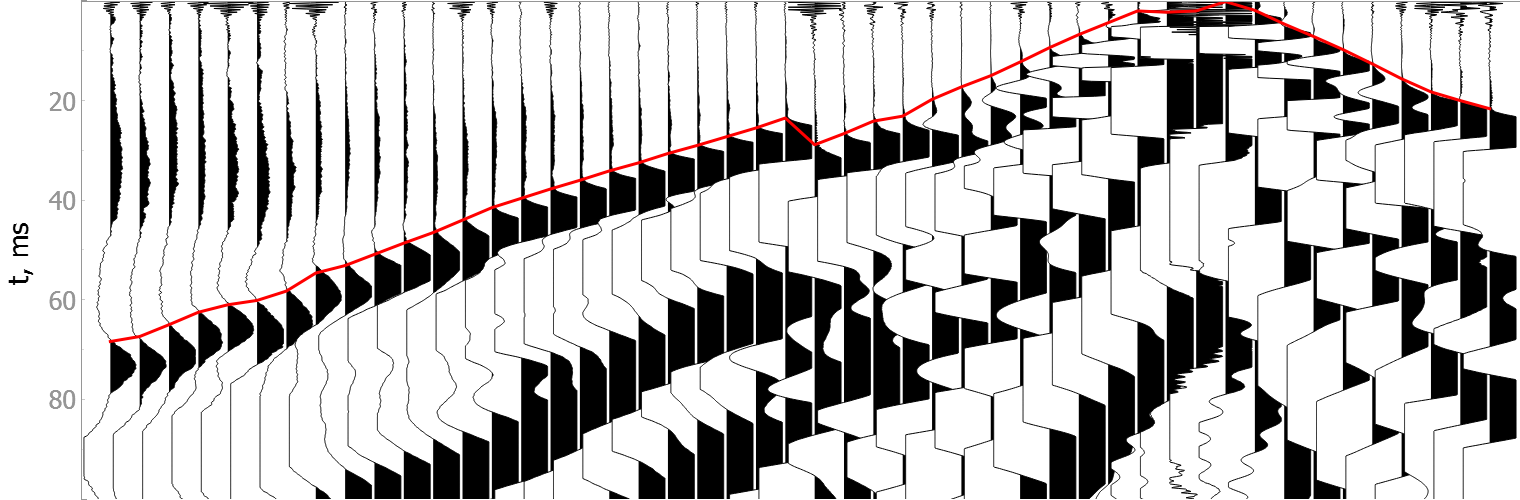Noisy data
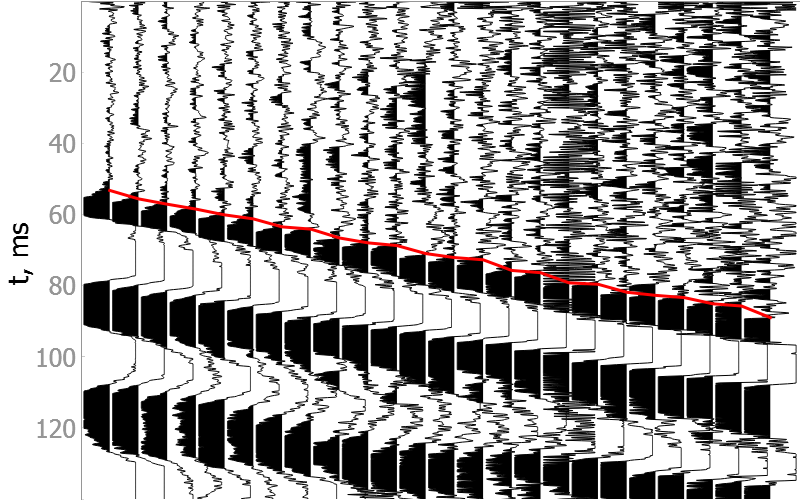 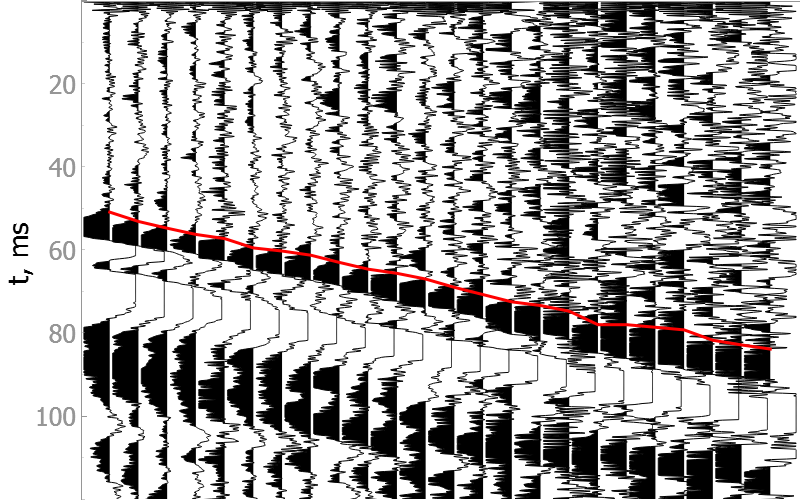     How model was trained?
Model training is described in detail here in the previous version of the project README.
The latest model was trained similarly, but: 1) Real land seismic data is used instead of synthetic data. 2) UNet3+ model is used, instead of the vanilla UNet.
Installation
Library is available in PyPI:
pip install -U first-breaks-pickingGPU support
You can use the capabilities of GPU (discrete, not integrated with CPU) to significantly reduce picking time. Before started, check here that your GPU is CUDA compatible.
Install GPU supported version of library:
pip install -U first-breaks-picking-gpuThe following steps are operating system dependent and must be performed manually:
- Install latest NVIDIA drivers.
- Install CUDA toolkit. The version must be between 11.x, starting with 11.6. Version 12 also may work, but versions >=11.6 are recommended.
- Install ZLib and CuDNN: Windows and Linux.
Compiled desktop application
Compiled executable files (.exe in .zip archives), as well as installers (.msi), can be downloaded in the Releases section.
If you want to use the capabilities of your dedicated GPU, also follow the steps in the previous step to install the driver, CUDA, CuDNN, and ZLib.
You can run the desktop application on other operating systems (Linux, MacOS) as well, but it must be run using the Python environment.
Extra data
-
To pick first breaks you need to download model.
-
If you have no seismic data, you can also download small SGY file.
It's also possible to download them with Python using the following snippet:
from first_breaks.utils.utils import (download_demo_sgy,
download_model_onnx)
sgy_filename = 'data.sgy'
model_filename = 'model.onnx'
download_demo_sgy(sgy_filename)
download_model_onnx(model_filename)How to use it
The library can be used in Python, or you can use the desktop application.
Python
Programmatic way has more flexibility for building your own picking scenario and processing multiple files.
Minimal example
The following snippet implements the picking process of the demo file. As a result, you can get an image from the project preview.
from first_breaks.utils.utils import download_demo_sgy
from first_breaks.sgy.reader import SGY
from first_breaks.picking.task import Task
from first_breaks.picking.picker_onnx import PickerONNX
from first_breaks.desktop.graph import export_image
sgy_filename = 'data.sgy'
download_demo_sgy(fname=sgy_filename)
sgy = SGY(sgy_filename)
task = Task(source=sgy,
traces_per_gather=12,
maximum_time=100,
gain=2)
picker = PickerONNX()
task = picker.process_task(task)
task.picks.color = (255, 0, 0)
# create an image from the project preview
image_filename = 'project_preview.png'
export_image(task, image_filename,
time_window=(0, 60),
traces_window=(79.5, 90.5),
show_processing_region=False,
headers_total_pixels=80,
height=500,
width=700,
hide_traces_axis=True
)For a better understanding of the steps taken, expand and read the next section.
Detailed examples
Show examples
### Download demo SGY Let's download the demo file. All the following examples assume that the file is downloaded and saved as `data.sgy`. You can also put your own SGY file. ```python from first_breaks.utils.utils import download_demo_sgy sgy_filename = 'data.sgy' download_demo_sgy(fname=sgy_filename) ``` ### Create SGY We provide several ways to create `SGY` object: from file, `bytes` or `numpy` array. From file: [code-block-start]:init-from-path ```python from first_breaks.sgy.reader import SGY sgy_filename = 'data.sgy' sgy = SGY(sgy_filename) ``` [code-block-end]:init-from-path From `bytes`: [code-block-start]:init-from-bytes ```python from first_breaks.sgy.reader import SGY sgy_filename = 'data.sgy' with open(sgy_filename, 'rb') as fin: sgy_bytes = fin.read() sgy = SGY(sgy_bytes) ``` [code-block-end]:init-from-bytes If you want to create from `numpy` array, extra argument `dt_mcs` is required: [code-block-start]:init-from-np ```python import numpy as np from first_breaks.sgy.reader import SGY num_samples = 1000 num_traces = 48 dt_mcs = 1e3 traces = np.random.random((num_samples, num_traces)) sgy = SGY(traces, dt_mcs=dt_mcs) ``` [code-block-end]:init-from-np ### Content of SGY Created `SGY` allows you to read traces, get observation parameters and view headers (empty if created from `numpy`) [code-block-start]:sgy-content ```python from first_breaks.sgy.reader import SGY sgy_filename = 'data.sgy' sgy = SGY(sgy_filename) # get all traces or specific traces limited by time all_traces = sgy.read() block_of_data = sgy.read_traces_by_ids(ids=[1, 2, 3, 10], min_sample=100, max_sample=500) # number of traces, values are the same print(sgy.num_traces, sgy.ntr) # number of time samples, values are the same print(sgy.num_samples, sgy.ns) # = (ns, ntr) print(sgy.shape) # time discretization, in mcs, in mcs, in ms, fs in Hertz print(sgy.dt, sgy.dt_mcs, sgy.dt_ms, sgy.fs) # dict with headers in the first 3600 bytes of the file print(sgy.general_headers) # pandas DataFrame with headers for each trace print(sgy.traces_headers.head()) ``` [code-block-end]:sgy-content ### Create task for picking Next, we create a task for picking and pass the picking parameters to it. They have default values, but for the best quality, they must be matched to specific data. You can use the desktop application to evaluate the parameters. A detailed description of the parameters can be found in the `Picking process` chapter. [code-block-start]:create-task ```python from first_breaks.sgy.reader import SGY from first_breaks.picking.task import Task sgy_filename = 'data.sgy' sgy = SGY(sgy_filename) task = Task(source=sgy, traces_per_gather=24, maximum_time=200) ``` [code-block-end]:create-task ### Create Picker In this step, we instantiate the neural network for picking. If you downloaded the model according to the installation section, then pass the path to it. Or leave the path to the model empty so that we can download it automatically. It's also possible to use GPU/CUDA to accelerate computation. By default `cuda` is selected if you have finished all steps regarding GPU in `Installation` section. Otherwise, it's `cpu`. You can also set the value of parameter `batch_size`, which can further speed up the calculations on the GPU. However, this will require additional video memory (VRAM). NOTE: When using the CPU, increasing `batch_size` does not speed up the calculation at all, but it may require additional memory costs (RAM). So don't increase this parameter when using CPU. [code-block-start]:create-picker ```python from first_breaks.picking.picker_onnx import PickerONNX # the library will determine the best available device picker_default = PickerONNX() # create picker explicitly on CPU picker_cpu = PickerONNX(device='cpu') # create picker explicitly on GPU picker_gpu = PickerONNX(device='cuda', batch_size=2) # transfer model to another device picker_cpu.change_settings(device='cuda', batch_size=3) picker_gpu.change_settings(device='cpu', batch_size=1) ``` [code-block-end]:create-picker ### Pick first breaks Now, using all the created components, we can pick the first breaks and retrieve results. [code-block-start]:pick-fb ```python from first_breaks.picking.task import Task from first_breaks.picking.picker_onnx import PickerONNX from first_breaks.sgy.reader import SGY sgy_filename = 'data.sgy' sgy = SGY(sgy_filename) task = Task(source=sgy, traces_per_gather=24, maximum_time=200) picker = PickerONNX() task = picker.process_task(task) # picks available from attribute picks = task.picks # or by method picks = task.get_result() # print values and confidence of picks print(picks.picks_in_mcs, picks.confidence) # print picking parameters print(picks.picking_parameters, task.picking_parameters) ``` [code-block-end]:pick-fb ### Picks The picks are stored in class `Picks`. In addition to the pick values, the class stores additional information (picking parameters, color, etc.). The class instance is returned as a picking result, but can also be created manually. [code-block-start]:picks ```python from first_breaks.picking.picks import Picks picks_in_samples = [1, 10, 20, 30, 50, 100] dt_mcs = 250 picks = Picks(values=picks_in_samples, unit="mcs", dt_mcs=dt_mcs) # get values print(picks.picks_in_samples) print(picks.picks_in_mcs) print(picks.picks_in_ms) # change color and width for visualisation picks.color = (255, 0, 0) picks.width = 5 # if picks are obtained by NN you can read extra attributes print(picks.confidence) print(picks.picking_parameters) print(picks.created_by_nn) ``` [code-block-end]:picks ### Visualizations You can save the seismogram and picks as an image. We use Qt backend for visualizations. Here we describe some usage scenarios. We've added named arguments to various scenarios for demonstration purposes, but in practice you can use them all. See the function arguments for more visualization options. Plot `SGY` only: [code-block-start]:plot-sgy ```python from first_breaks.sgy.reader import SGY from first_breaks.desktop.graph import export_image sgy_filename = 'data.sgy' image_filename = 'image.png' sgy = SGY(sgy_filename) export_image(sgy, image_filename, normalize=None, traces_window=(5, 10), time_window=(0, 200), height=300, width_per_trace=30) ``` [code-block-end]:plot-sgy Plot `numpy` traces: [code-block-start]:plot-np ```python import numpy as np from first_breaks.sgy.reader import SGY from first_breaks.desktop.graph import export_image image_filename = 'image.png' num_traces = 48 num_samples = 1000 dt_mcs = 1e3 traces = np.random.random((num_samples, num_traces)) export_image(traces, image_filename, dt_mcs=dt_mcs, clip=0.5) # or create SGY as discussed before sgy = SGY(traces, dt_mcs=dt_mcs) export_image(sgy, image_filename, gain=2) ``` [code-block-end]:plot-np Plot `SGY` with custom picks: [code-block-start]:plot-sgy-custom-picks ```python import numpy as np from first_breaks.picking.picks import Picks from first_breaks.sgy.reader import SGY from first_breaks.desktop.graph import export_image sgy_filename = 'data.sgy' image_filename = 'image.png' sgy = SGY(sgy_filename) picks_ms = np.random.uniform(low=0, high=sgy.ns * sgy.dt_ms, size=sgy.ntr) picks = Picks(values=picks_ms, unit="ms", dt_mcs=sgy.dt_mcs, color=(0, 100, 100)) export_image(sgy, image_filename, picks=picks) ``` [code-block-end]:plot-sgy-custom-picks Plot result of picking: [code-block-start]:plot-sgy-real-picks ```python from first_breaks.picking.task import Task from first_breaks.picking.picker_onnx import PickerONNX from first_breaks.desktop.graph import export_image from first_breaks.sgy.reader import SGY sgy_filename = 'data.sgy' image_filename = 'image.png' sgy = SGY(sgy_filename) task = Task(source=sgy, traces_per_gather=24, maximum_time=200) picker = PickerONNX() task = picker.process_task(task) export_image(task, image_filename, show_processing_region=False, fill_black='right', width=1000) ``` [code-block-end]:plot-sgy-real-picks ### *Limit processing region Unfortunately, processing of a part of a file is not currently supported natively. We will add this functionality soon! However, you can use the following workaround to do this: [code-block-start]:pick-limited ```python from first_breaks.sgy.reader import SGY sgy_filename = 'data.sgy' sgy = SGY(sgy_filename) interesting_traces = sgy.read_traces_by_ids(ids=list(range(20, 40)), min_sample=100, max_sample=200) # we create new SGY based on region of interests sgy = SGY(interesting_traces, dt_mcs=sgy.dt_mcs) ``` [code-block-end]:pick-limitedDesktop application
Application under development
Desktop application allows you to work interactively with only one file and has better performance in visualization. You can use application as SGY viewer, as well as visually evaluate the optimal values of the picking parameters for your data.
Launch app
Enter command to launch the application
first-breaks-picking appor
first-breaks-picking desktopSelect and view SGY file
Click on 2 button to select SGY. After successful reading you can analyze SGY file.
The following mouse interactions are available:
- Left button drag / Middle button drag: Pan the scene.
- Right button drag: Scales the scene. Dragging left/right scales horizontally; dragging up/down scales vertically.
- Right button click: Open dialog with extra options, such as limit by X/Y axes and export.
- Wheel spin: Zooms the scene in and out.
- Left click: You can manually change values of active picks.
Spectrum analysis:
- Hold down the Shift key, the left mouse button, and select an area: a window with a spectrum will appear. You can create as many windows as you like.
- Move existing windows by dragging with the left mouse button.
- Delete the window with the spectrum by clicking on it and pressing the Del key.
Load model
To use picker in desktop app you have to download model. See the Installation section for instructions
on how to download the model.
Click on 1 button and select file with model. After successfully loading the model, access to the pick with NN will open.
Settings and Processing
Click on 3 button to open window with different settings and picking parameters:
- The Processing section contains parameters for drawing seismograms; these parameters are also used during the picking process. Changing them in real time changes the rendering.
- The View section allows you to change the axes' names, content, and orientation.
- The NN Picking section allows you to select additional parameters for picking the first arrivals and a device
for calculations. If you have CUDA compatible GPU and installed GPU supported version of library
(see
Installationsection), you can selectCUDA/GPUdevice to use GPU acceleration. It can drastically decrease computation time.
A detailed description of the parameters responsible for picking can be found in the Picking process chapter.
When you have chosen the optimal parameters, click on the Run picking button. After some time, a line will appear connecting the first arrivals. You can interrupt the process by clicking Stop button.
Processing grid
Click on 4 button to toggle the display of the processing grid on or off. Horizontal line
shows Maximum time and vertical lines are drawn at intervals equal to Traces per gather. The neural network
processes blocks independently, as separate images.
Picks manager
Click on 5 button to open picks manager.
When picking with a neural network, the result of the picking will appear here. You can also: add manual picks, load picks from headers, duplicate picks, aggregate several picks, or remove picks.
As control and navigation tools, you can choose to show picks or not (checkbox), select active picks (radio button), and change color (color label).
After selecting one of the picks, you can double-click or click the button with the “save” icon to show the export options.
Picking process
Neural network process file as series of images. There is why the traces should not be random, since we are using information about adjacent traces.
To obtain the first breaks we do the following steps:
1) Read all traces in the file.
 2) Limit time range by
2) Limit time range by Maximum time.
 3) Split the sequence of traces into independent gathers of lengths
3) Split the sequence of traces into independent gathers of lengths Traces per gather each.
 4) Apply trace modification on the gathers level if necessary (
4) Apply trace modification on the gathers level if necessary (Gain, Clip, etc).
5) Calculate first breaks for individual gathers independently.
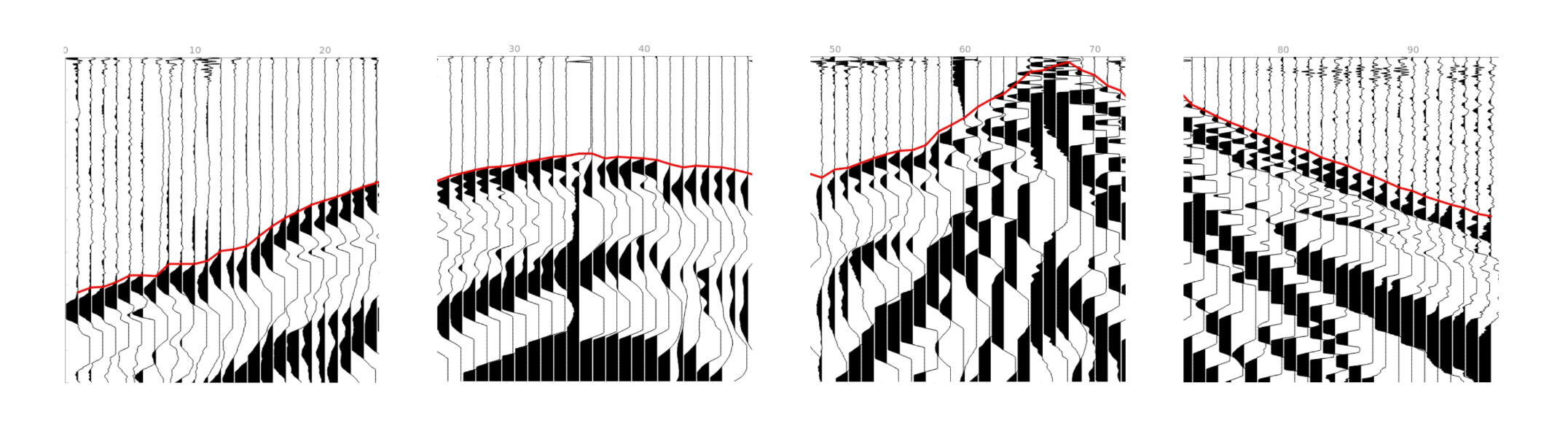 6) Join the first breaks of individual gathers.
6) Join the first breaks of individual gathers.
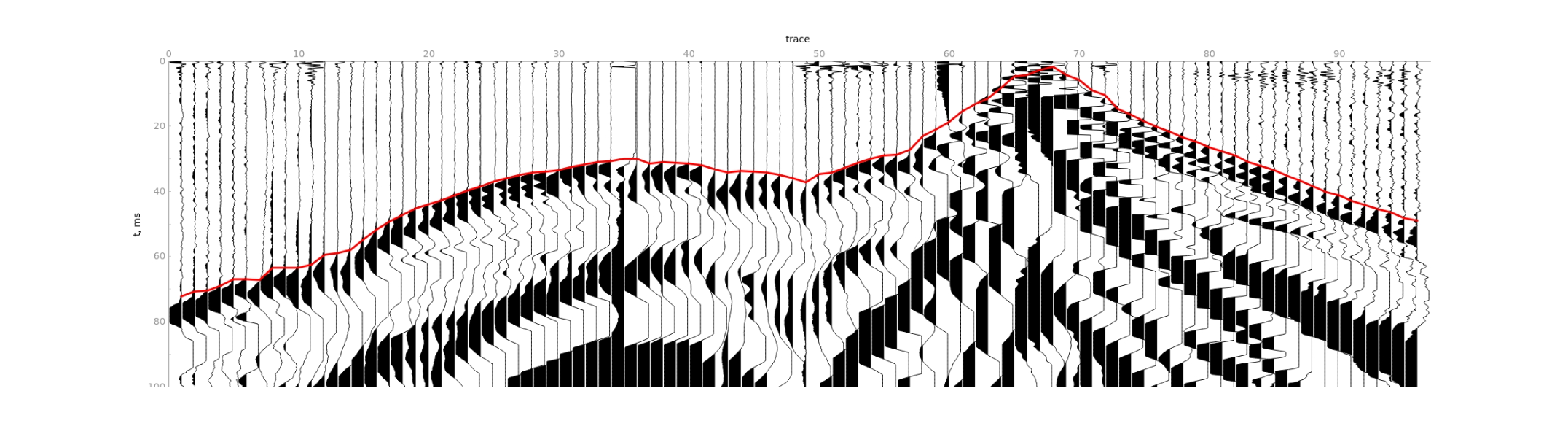
To achieve the best result, you need to modify the picking parameters.
Traces per gather
Traces per gather is the most important parameter for picking. The parameter defines how we split the sequence of
traces into individual gathers.
Suppose we need to process a file with 96 traces. Depending on the value of Traces per gather parameter, we will
process it as follows:
Traces per gather= 24. We will process 4 gathers with 24 traces each.
Traces per gather= 48. We will process 2 gathers with 48 traces each.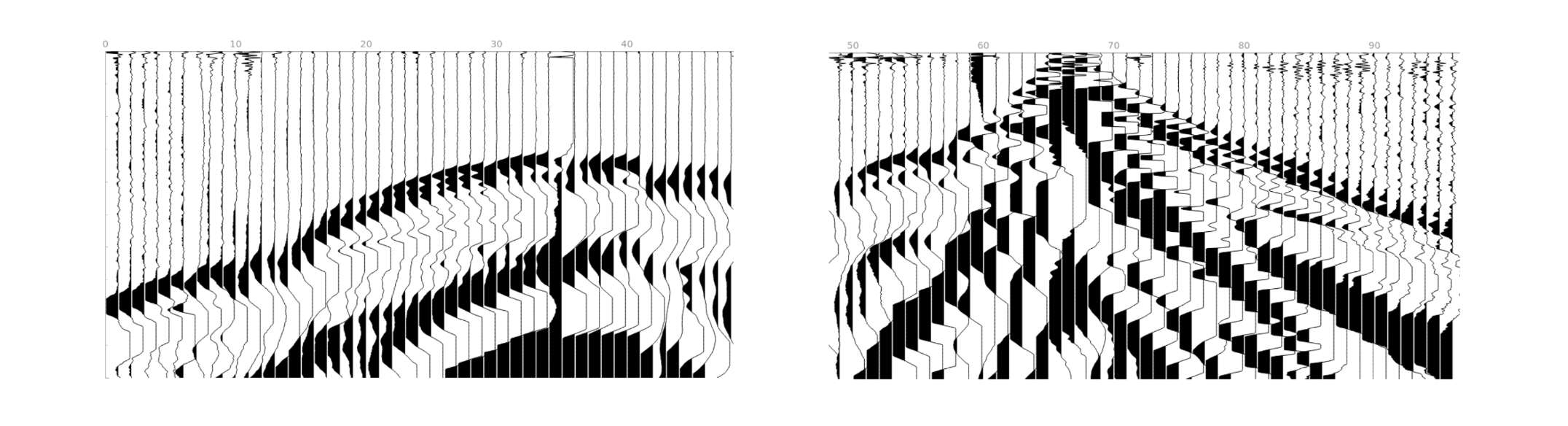
Traces per gather= 40. We will process 2 gathers with 40 traces each and 1 gather with the remaining 16 traces. The last gather will be interpolated from 16 to 40 traces.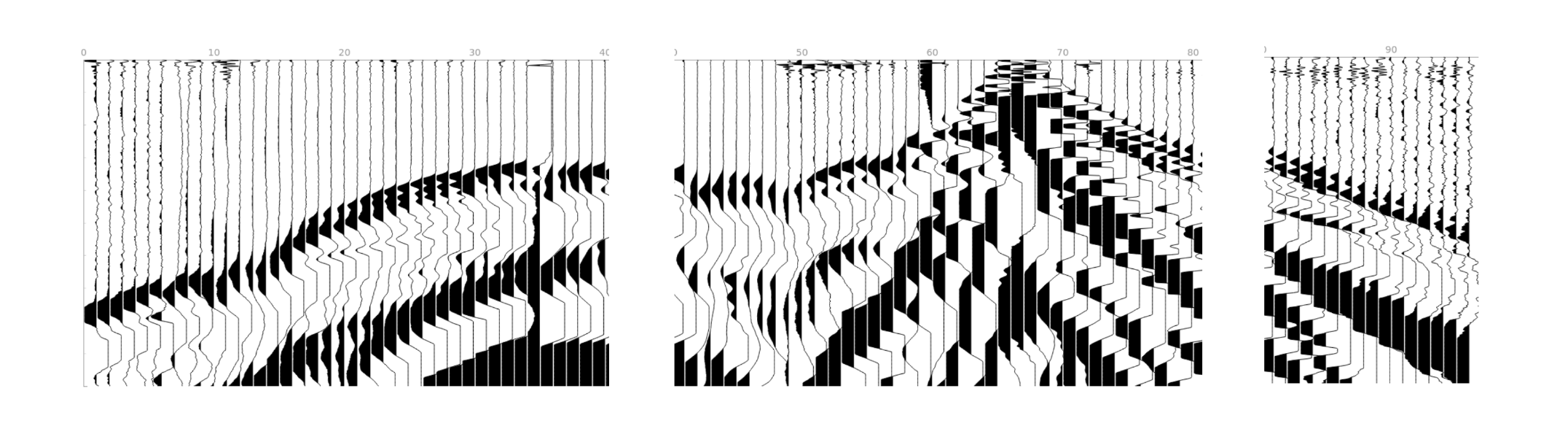
Maximum time
You can localize the area for finding first breaks. Specify Maximum time if you have long records but the first breaks
located at the start of the traces. Keep it 0 if you don't want to limit traces.
List of traces to inverse
Some receivers may have the wrong polarity, so you can specify which traces should be inversed. Note, that inversion
will be applied on the gathers level. For example, if you have 96 traces, Traces per gather = 48 and
List of traces to inverse = (2, 30, 48), then traces (2, 30, 48, 50, 78, 96) will be inversed.
Notes:
- Trace indexing starts at 1.
- Option is not available on desktop app.
Recommendations
You can receive predictions for any file with any parameters, but to get a better result, you should comply with the following guidelines:
- Your file should contain one or more gathers. By a gather, we mean that traces within a single gather can be geophysically interpreted. The traces within the same gather should not be random, since we are using information about adjacent traces.
- All gathers in the file must contain the same number of traces.
- The number of traces in gather must be equal to
Traces per gatheror divisible by it without a remainder. For example, if you have CSP gathers and the number of receivers is 48, then you can set the parameter value to 48, 24, or 12. - We don't sort your file (CMP, CRP, CSP, etc), so you should send us files with traces sorted by yourself.
- You can process a file with independent seismograms obtained from different polygons, under different conditions, etc., but the requirements listed above must be met.
Acknowledgments
We would like to thank GEODEVICE for providing field data from land and borehole seismic surveys with annotated first breaks for model training.
