🚀 LightRAG: Simple and Fast Retrieval-Augmented Generation

🎉 News
- [x] [2024.11.19]🎯📢We have added a detailed blog link introducing LightRAG on LearnOpenCV. Many thanks to the blog author!
- [x] [2024.11.12]🎯📢You can use Oracle Database 23ai for all storage types (kv/vector/graph) now.
- [x] [2024.11.11]🎯📢LightRAG now supports deleting entities by their names.
- [x] [2024.11.09]🎯📢Introducing the LightRAG Gui, which allows you to insert, query, visualize, and download LightRAG knowledge.
- [x] [2024.11.04]🎯📢You can now use Neo4J for Storage.
- [x] [2024.10.29]🎯📢LightRAG now supports multiple file types, including PDF, DOC, PPT, and CSV via
textract. - [x] [2024.10.20]🎯📢We’ve added a new feature to LightRAG: Graph Visualization.
- [x] [2024.10.18]🎯📢We’ve added a link to a LightRAG Introduction Video. Thanks to the author!
- [x] [2024.10.17]🎯📢We have created a Discord channel! Welcome to join for sharing and discussions! 🎉🎉
- [x] [2024.10.16]🎯📢LightRAG now supports Ollama models!
- [x] [2024.10.15]🎯📢LightRAG now supports Hugging Face models!
Algorithm Flowchart
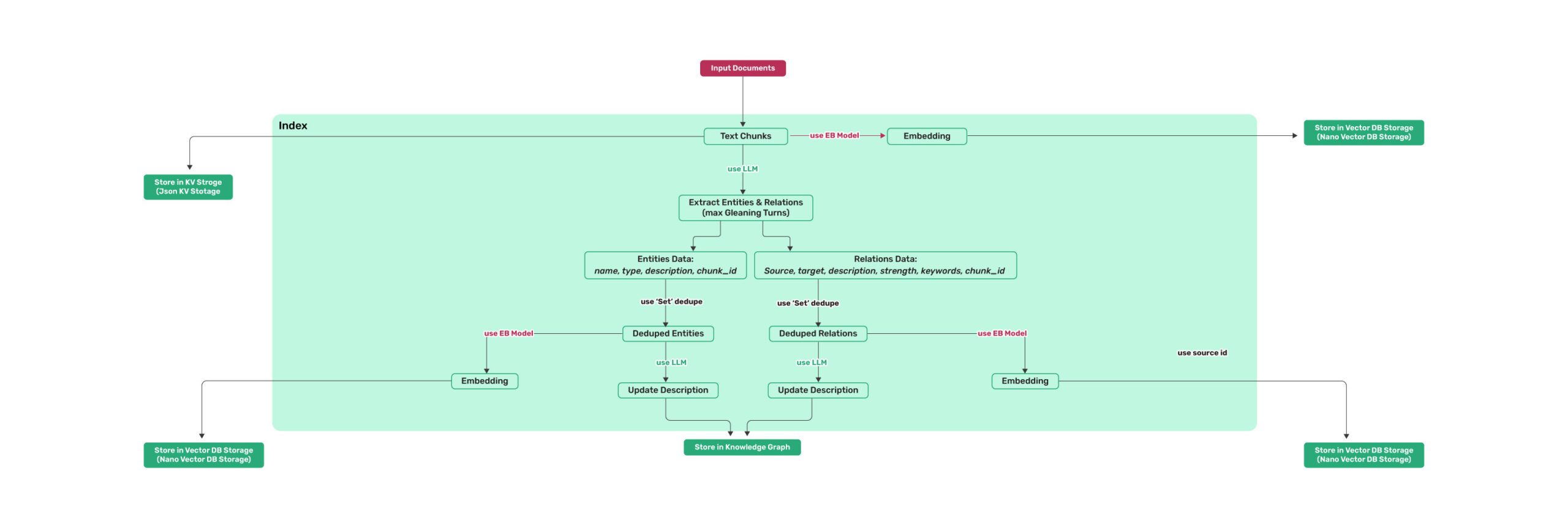 Figure 1: LightRAG Indexing Flowchart
Figure 1: LightRAG Indexing Flowchart
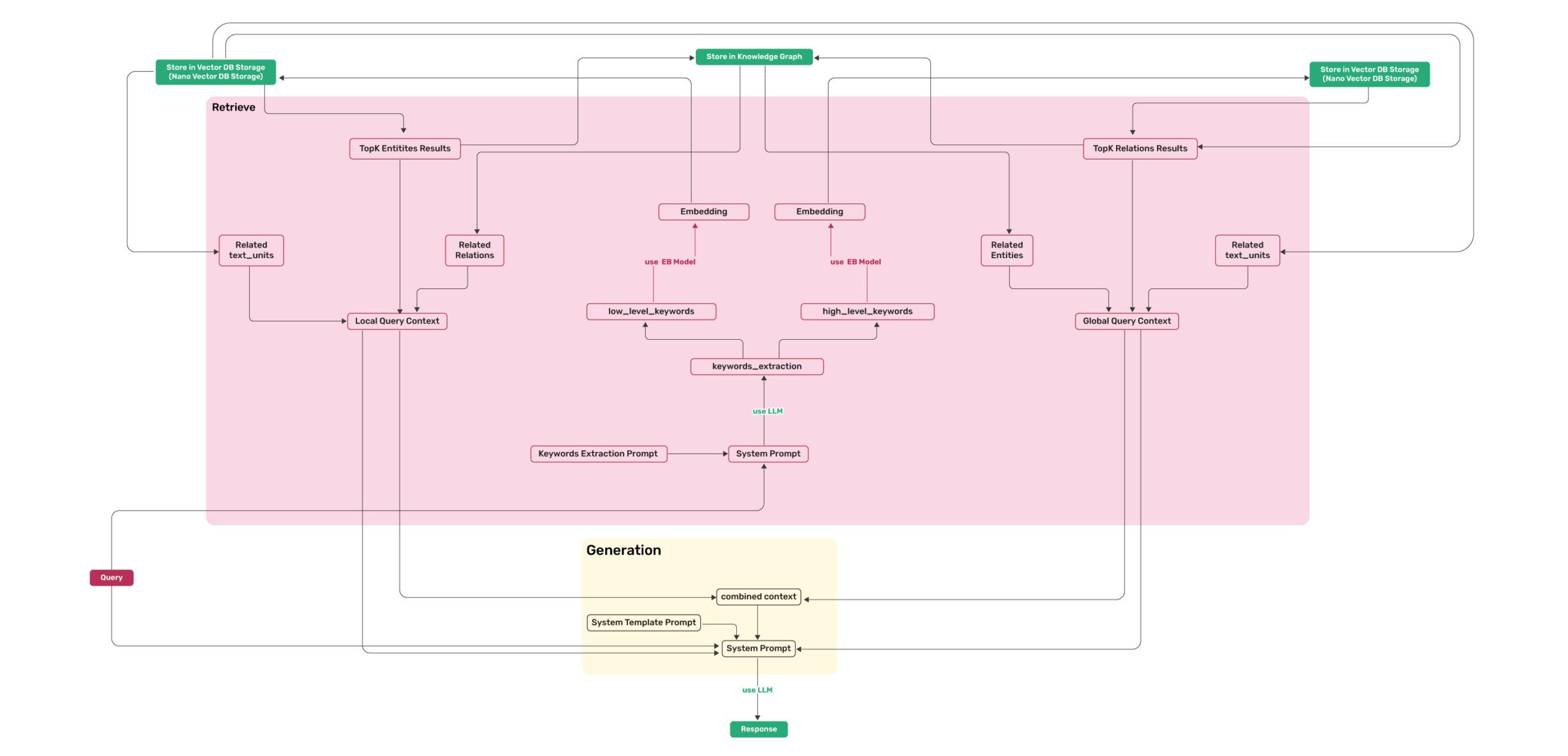 Figure 2: LightRAG Retrieval and Querying Flowchart
Figure 2: LightRAG Retrieval and Querying Flowchart
Install
- Install from source (Recommend)
cd LightRAG
pip install -e .- Install from PyPI
pip install lightrag-hku
Quick Start
- Video demo of running LightRAG locally.
- All the code can be found in the
examples. - Set OpenAI API key in environment if using OpenAI models:
export OPENAI_API_KEY="sk-...". - Download the demo text "A Christmas Carol by Charles Dickens":
curl https://raw.githubusercontent.com/gusye1234/nano-graphrag/main/tests/mock_data.txt > ./book.txtUse the below Python snippet (in a script) to initialize LightRAG and perform queries:
import os
from lightrag import LightRAG, QueryParam
from lightrag.llm import gpt_4o_mini_complete, gpt_4o_complete
#########
# Uncomment the below two lines if running in a jupyter notebook to handle the async nature of rag.insert()
# import nest_asyncio
# nest_asyncio.apply()
#########
WORKING_DIR = "./dickens"
if not os.path.exists(WORKING_DIR):
os.mkdir(WORKING_DIR)
rag = LightRAG(
working_dir=WORKING_DIR,
llm_model_func=gpt_4o_mini_complete # Use gpt_4o_mini_complete LLM model
# llm_model_func=gpt_4o_complete # Optionally, use a stronger model
)
with open("./book.txt") as f:
rag.insert(f.read())
# Perform naive search
print(rag.query("What are the top themes in this story?", param=QueryParam(mode="naive")))
# Perform local search
print(rag.query("What are the top themes in this story?", param=QueryParam(mode="local")))
# Perform global search
print(rag.query("What are the top themes in this story?", param=QueryParam(mode="global")))
# Perform hybrid search
print(rag.query("What are the top themes in this story?", param=QueryParam(mode="hybrid")))Using Open AI-like APIs
* LightRAG also supports Open AI-like chat/embeddings APIs: ```python async def llm_model_func( prompt, system_prompt=None, history_messages=[], **kwargs ) -> str: return await openai_complete_if_cache( "solar-mini", prompt, system_prompt=system_prompt, history_messages=history_messages, api_key=os.getenv("UPSTAGE_API_KEY"), base_url="https://api.upstage.ai/v1/solar", **kwargs ) async def embedding_func(texts: list[str]) -> np.ndarray: return await openai_embedding( texts, model="solar-embedding-1-large-query", api_key=os.getenv("UPSTAGE_API_KEY"), base_url="https://api.upstage.ai/v1/solar" ) rag = LightRAG( working_dir=WORKING_DIR, llm_model_func=llm_model_func, embedding_func=EmbeddingFunc( embedding_dim=4096, max_token_size=8192, func=embedding_func ) ) ```Using Hugging Face Models
* If you want to use Hugging Face models, you only need to set LightRAG as follows: ```python from lightrag.llm import hf_model_complete, hf_embedding from transformers import AutoModel, AutoTokenizer from lightrag.utils import EmbeddingFunc # Initialize LightRAG with Hugging Face model rag = LightRAG( working_dir=WORKING_DIR, llm_model_func=hf_model_complete, # Use Hugging Face model for text generation llm_model_name='meta-llama/Llama-3.1-8B-Instruct', # Model name from Hugging Face # Use Hugging Face embedding function embedding_func=EmbeddingFunc( embedding_dim=384, max_token_size=5000, func=lambda texts: hf_embedding( texts, tokenizer=AutoTokenizer.from_pretrained("sentence-transformers/all-MiniLM-L6-v2"), embed_model=AutoModel.from_pretrained("sentence-transformers/all-MiniLM-L6-v2") ) ), ) ```Using Ollama Models
### Overview If you want to use Ollama models, you need to pull model you plan to use and embedding model, for example `nomic-embed-text`. Then you only need to set LightRAG as follows: ```python from lightrag.llm import ollama_model_complete, ollama_embedding from lightrag.utils import EmbeddingFunc # Initialize LightRAG with Ollama model rag = LightRAG( working_dir=WORKING_DIR, llm_model_func=ollama_model_complete, # Use Ollama model for text generation llm_model_name='your_model_name', # Your model name # Use Ollama embedding function embedding_func=EmbeddingFunc( embedding_dim=768, max_token_size=8192, func=lambda texts: ollama_embedding( texts, embed_model="nomic-embed-text" ) ), ) ``` ### Using Neo4J for Storage * For production level scenarios you will most likely want to leverage an enterprise solution * for KG storage. Running Neo4J in Docker is recommended for seamless local testing. * See: https://hub.docker.com/_/neo4j ```python export NEO4J_URI="neo4j://localhost:7687" export NEO4J_USERNAME="neo4j" export NEO4J_PASSWORD="password" When you launch the project be sure to override the default KG: NetworkS by specifying kg="Neo4JStorage". # Note: Default settings use NetworkX #Initialize LightRAG with Neo4J implementation. WORKING_DIR = "./local_neo4jWorkDir" rag = LightRAG( working_dir=WORKING_DIR, llm_model_func=gpt_4o_mini_complete, # Use gpt_4o_mini_complete LLM model kg="Neo4JStorage", #<-----------override KG default log_level="DEBUG" #<-----------override log_level default ) ``` see test_neo4j.py for a working example. ### Increasing context size In order for LightRAG to work context should be at least 32k tokens. By default Ollama models have context size of 8k. You can achieve this using one of two ways: #### Increasing the `num_ctx` parameter in Modelfile. 1. Pull the model: ```bash ollama pull qwen2 ``` 2. Display the model file: ```bash ollama show --modelfile qwen2 > Modelfile ``` 3. Edit the Modelfile by adding the following line: ```bash PARAMETER num_ctx 32768 ``` 4. Create the modified model: ```bash ollama create -f Modelfile qwen2m ``` #### Setup `num_ctx` via Ollama API. Tiy can use `llm_model_kwargs` param to configure ollama: ```python rag = LightRAG( working_dir=WORKING_DIR, llm_model_func=ollama_model_complete, # Use Ollama model for text generation llm_model_name='your_model_name', # Your model name llm_model_kwargs={"options": {"num_ctx": 32768}}, # Use Ollama embedding function embedding_func=EmbeddingFunc( embedding_dim=768, max_token_size=8192, func=lambda texts: ollama_embedding( texts, embed_model="nomic-embed-text" ) ), ) ``` #### Fully functional example There fully functional example `examples/lightrag_ollama_demo.py` that utilizes `gemma2:2b` model, runs only 4 requests in parallel and set context size to 32k. #### Low RAM GPUs In order to run this experiment on low RAM GPU you should select small model and tune context window (increasing context increase memory consumption). For example, running this ollama example on repurposed mining GPU with 6Gb of RAM required to set context size to 26k while using `gemma2:2b`. It was able to find 197 entities and 19 relations on `book.txt`.Query Param
class QueryParam:
mode: Literal["local", "global", "hybrid", "naive"] = "global"
only_need_context: bool = False
response_type: str = "Multiple Paragraphs"
# Number of top-k items to retrieve; corresponds to entities in "local" mode and relationships in "global" mode.
top_k: int = 60
# Number of tokens for the original chunks.
max_token_for_text_unit: int = 4000
# Number of tokens for the relationship descriptions
max_token_for_global_context: int = 4000
# Number of tokens for the entity descriptions
max_token_for_local_context: int = 4000Batch Insert
# Batch Insert: Insert multiple texts at once
rag.insert(["TEXT1", "TEXT2",...])Incremental Insert
# Incremental Insert: Insert new documents into an existing LightRAG instance
rag = LightRAG(
working_dir=WORKING_DIR,
llm_model_func=llm_model_func,
embedding_func=EmbeddingFunc(
embedding_dim=embedding_dimension,
max_token_size=8192,
func=embedding_func,
),
)
with open("./newText.txt") as f:
rag.insert(f.read())Delete Entity
# Delete Entity: Deleting entities by their names
rag = LightRAG(
working_dir=WORKING_DIR,
llm_model_func=llm_model_func,
embedding_func=EmbeddingFunc(
embedding_dim=embedding_dimension,
max_token_size=8192,
func=embedding_func,
),
)
rag.delete_by_entity("Project Gutenberg")Multi-file Type Support
The textract supports reading file types such as TXT, DOCX, PPTX, CSV, and PDF.
import textract
file_path = 'TEXT.pdf'
text_content = textract.process(file_path)
rag.insert(text_content.decode('utf-8'))Graph Visualization
Graph visualization with html
* The following code can be found in `examples/graph_visual_with_html.py` ```python import networkx as nx from pyvis.network import Network # Load the GraphML file G = nx.read_graphml('./dickens/graph_chunk_entity_relation.graphml') # Create a Pyvis network net = Network(notebook=True) # Convert NetworkX graph to Pyvis network net.from_nx(G) # Save and display the network net.show('knowledge_graph.html') ```Graph visualization with Neo4j
* The following code can be found in `examples/graph_visual_with_neo4j.py` ```python import os import json from lightrag.utils import xml_to_json from neo4j import GraphDatabase # Constants WORKING_DIR = "./dickens" BATCH_SIZE_NODES = 500 BATCH_SIZE_EDGES = 100 # Neo4j connection credentials NEO4J_URI = "bolt://localhost:7687" NEO4J_USERNAME = "neo4j" NEO4J_PASSWORD = "your_password" def convert_xml_to_json(xml_path, output_path): """Converts XML file to JSON and saves the output.""" if not os.path.exists(xml_path): print(f"Error: File not found - {xml_path}") return None json_data = xml_to_json(xml_path) if json_data: with open(output_path, 'w', encoding='utf-8') as f: json.dump(json_data, f, ensure_ascii=False, indent=2) print(f"JSON file created: {output_path}") return json_data else: print("Failed to create JSON data") return None def process_in_batches(tx, query, data, batch_size): """Process data in batches and execute the given query.""" for i in range(0, len(data), batch_size): batch = data[i:i + batch_size] tx.run(query, {"nodes": batch} if "nodes" in query else {"edges": batch}) def main(): # Paths xml_file = os.path.join(WORKING_DIR, 'graph_chunk_entity_relation.graphml') json_file = os.path.join(WORKING_DIR, 'graph_data.json') # Convert XML to JSON json_data = convert_xml_to_json(xml_file, json_file) if json_data is None: return # Load nodes and edges nodes = json_data.get('nodes', []) edges = json_data.get('edges', []) # Neo4j queries create_nodes_query = """ UNWIND $nodes AS node MERGE (e:Entity {id: node.id}) SET e.entity_type = node.entity_type, e.description = node.description, e.source_id = node.source_id, e.displayName = node.id REMOVE e:Entity WITH e, node CALL apoc.create.addLabels(e, [node.entity_type]) YIELD node AS labeledNode RETURN count(*) """ create_edges_query = """ UNWIND $edges AS edge MATCH (source {id: edge.source}) MATCH (target {id: edge.target}) WITH source, target, edge, CASE WHEN edge.keywords CONTAINS 'lead' THEN 'lead' WHEN edge.keywords CONTAINS 'participate' THEN 'participate' WHEN edge.keywords CONTAINS 'uses' THEN 'uses' WHEN edge.keywords CONTAINS 'located' THEN 'located' WHEN edge.keywords CONTAINS 'occurs' THEN 'occurs' ELSE REPLACE(SPLIT(edge.keywords, ',')[0], '\"', '') END AS relType CALL apoc.create.relationship(source, relType, { weight: edge.weight, description: edge.description, keywords: edge.keywords, source_id: edge.source_id }, target) YIELD rel RETURN count(*) """ set_displayname_and_labels_query = """ MATCH (n) SET n.displayName = n.id WITH n CALL apoc.create.setLabels(n, [n.entity_type]) YIELD node RETURN count(*) """ # Create a Neo4j driver driver = GraphDatabase.driver(NEO4J_URI, auth=(NEO4J_USERNAME, NEO4J_PASSWORD)) try: # Execute queries in batches with driver.session() as session: # Insert nodes in batches session.execute_write(process_in_batches, create_nodes_query, nodes, BATCH_SIZE_NODES) # Insert edges in batches session.execute_write(process_in_batches, create_edges_query, edges, BATCH_SIZE_EDGES) # Set displayName and labels session.run(set_displayname_and_labels_query) except Exception as e: print(f"Error occurred: {e}") finally: driver.close() if __name__ == "__main__": main() ```API Server Implementation
LightRAG also provides a FastAPI-based server implementation for RESTful API access to RAG operations. This allows you to run LightRAG as a service and interact with it through HTTP requests.
Setting up the API Server
Click to expand setup instructions
1. First, ensure you have the required dependencies: ```bash pip install fastapi uvicorn pydantic ``` 2. Set up your environment variables: ```bash export RAG_DIR="your_index_directory" # Optional: Defaults to "index_default" export OPENAI_BASE_URL="Your OpenAI API base URL" # Optional: Defaults to "https://api.openai.com/v1" export OPENAI_API_KEY="Your OpenAI API key" # Required export LLM_MODEL="Your LLM model" # Optional: Defaults to "gpt-4o-mini" export EMBEDDING_MODEL="Your embedding model" # Optional: Defaults to "text-embedding-3-large" ``` 3. Run the API server: ```bash python examples/lightrag_api_openai_compatible_demo.py ``` The server will start on `http://0.0.0.0:8020`.API Endpoints
The API server provides the following endpoints:
1. Query Endpoint
Click to view Query endpoint details
- **URL:** `/query` - **Method:** POST - **Body:** ```json { "query": "Your question here", "mode": "hybrid", // Can be "naive", "local", "global", or "hybrid" "only_need_context": true // Optional: Defaults to false, if true, only the referenced context will be returned, otherwise the llm answer will be returned } ``` - **Example:** ```bash curl -X POST "http://127.0.0.1:8020/query" \ -H "Content-Type: application/json" \ -d '{"query": "What are the main themes?", "mode": "hybrid"}' ```2. Insert Text Endpoint
Click to view Insert Text endpoint details
- **URL:** `/insert` - **Method:** POST - **Body:** ```json { "text": "Your text content here" } ``` - **Example:** ```bash curl -X POST "http://127.0.0.1:8020/insert" \ -H "Content-Type: application/json" \ -d '{"text": "Content to be inserted into RAG"}' ```3. Insert File Endpoint
Click to view Insert File endpoint details
- **URL:** `/insert_file` - **Method:** POST - **Body:** ```json { "file_path": "path/to/your/file.txt" } ``` - **Example:** ```bash curl -X POST "http://127.0.0.1:8020/insert_file" \ -H "Content-Type: application/json" \ -d '{"file_path": "./book.txt"}' ```4. Health Check Endpoint
Click to view Health Check endpoint details
- **URL:** `/health` - **Method:** GET - **Example:** ```bash curl -X GET "http://127.0.0.1:8020/health" ```Configuration
The API server can be configured using environment variables:
RAG_DIR: Directory for storing the RAG index (default: "index_default")- API keys and base URLs should be configured in the code for your specific LLM and embedding model providers
Error Handling
Click to view error handling details
The API includes comprehensive error handling: - File not found errors (404) - Processing errors (500) - Supports multiple file encodings (UTF-8 and GBK)Evaluation
Dataset
The dataset used in LightRAG can be downloaded from TommyChien/UltraDomain.
Generate Query
LightRAG uses the following prompt to generate high-level queries, with the corresponding code in example/generate_query.py.
Prompt
```python Given the following description of a dataset: {description} Please identify 5 potential users who would engage with this dataset. For each user, list 5 tasks they would perform with this dataset. Then, for each (user, task) combination, generate 5 questions that require a high-level understanding of the entire dataset. Output the results in the following structure: - User 1: [user description] - Task 1: [task description] - Question 1: - Question 2: - Question 3: - Question 4: - Question 5: - Task 2: [task description] ... - Task 5: [task description] - User 2: [user description] ... - User 5: [user description] ... ```Batch Eval
To evaluate the performance of two RAG systems on high-level queries, LightRAG uses the following prompt, with the specific code available in example/batch_eval.py.
Prompt
```python ---Role--- You are an expert tasked with evaluating two answers to the same question based on three criteria: **Comprehensiveness**, **Diversity**, and **Empowerment**. ---Goal--- You will evaluate two answers to the same question based on three criteria: **Comprehensiveness**, **Diversity**, and **Empowerment**. - **Comprehensiveness**: How much detail does the answer provide to cover all aspects and details of the question? - **Diversity**: How varied and rich is the answer in providing different perspectives and insights on the question? - **Empowerment**: How well does the answer help the reader understand and make informed judgments about the topic? For each criterion, choose the better answer (either Answer 1 or Answer 2) and explain why. Then, select an overall winner based on these three categories. Here is the question: {query} Here are the two answers: **Answer 1:** {answer1} **Answer 2:** {answer2} Evaluate both answers using the three criteria listed above and provide detailed explanations for each criterion. Output your evaluation in the following JSON format: {{ "Comprehensiveness": {{ "Winner": "[Answer 1 or Answer 2]", "Explanation": "[Provide explanation here]" }}, "Empowerment": {{ "Winner": "[Answer 1 or Answer 2]", "Explanation": "[Provide explanation here]" }}, "Overall Winner": {{ "Winner": "[Answer 1 or Answer 2]", "Explanation": "[Summarize why this answer is the overall winner based on the three criteria]" }} }} ```Overall Performance Table
| Agriculture | CS | Legal | Mix | |||||
|---|---|---|---|---|---|---|---|---|
| NaiveRAG | LightRAG | NaiveRAG | LightRAG | NaiveRAG | LightRAG | NaiveRAG | LightRAG | |
| Comprehensiveness | 32.4% | 67.6% | 38.4% | 61.6% | 16.4% | 83.6% | 38.8% | 61.2% |
| Diversity | 23.6% | 76.4% | 38.0% | 62.0% | 13.6% | 86.4% | 32.4% | 67.6% |
| Empowerment | 32.4% | 67.6% | 38.8% | 61.2% | 16.4% | 83.6% | 42.8% | 57.2% |
| Overall | 32.4% | 67.6% | 38.8% | 61.2% | 15.2% | 84.8% | 40.0% | 60.0% |
| RQ-RAG | LightRAG | RQ-RAG | LightRAG | RQ-RAG | LightRAG | RQ-RAG | LightRAG | |
| Comprehensiveness | 31.6% | 68.4% | 38.8% | 61.2% | 15.2% | 84.8% | 39.2% | 60.8% |
| Diversity | 29.2% | 70.8% | 39.2% | 60.8% | 11.6% | 88.4% | 30.8% | 69.2% |
| Empowerment | 31.6% | 68.4% | 36.4% | 63.6% | 15.2% | 84.8% | 42.4% | 57.6% |
| Overall | 32.4% | 67.6% | 38.0% | 62.0% | 14.4% | 85.6% | 40.0% | 60.0% |
| HyDE | LightRAG | HyDE | LightRAG | HyDE | LightRAG | HyDE | LightRAG | |
| Comprehensiveness | 26.0% | 74.0% | 41.6% | 58.4% | 26.8% | 73.2% | 40.4% | 59.6% |
| Diversity | 24.0% | 76.0% | 38.8% | 61.2% | 20.0% | 80.0% | 32.4% | 67.6% |
| Empowerment | 25.2% | 74.8% | 40.8% | 59.2% | 26.0% | 74.0% | 46.0% | 54.0% |
| Overall | 24.8% | 75.2% | 41.6% | 58.4% | 26.4% | 73.6% | 42.4% | 57.6% |
| GraphRAG | LightRAG | GraphRAG | LightRAG | GraphRAG | LightRAG | GraphRAG | LightRAG | |
| Comprehensiveness | 45.6% | 54.4% | 48.4% | 51.6% | 48.4% | 51.6% | 50.4% | 49.6% |
| Diversity | 22.8% | 77.2% | 40.8% | 59.2% | 26.4% | 73.6% | 36.0% | 64.0% |
| Empowerment | 41.2% | 58.8% | 45.2% | 54.8% | 43.6% | 56.4% | 50.8% | 49.2% |
| Overall | 45.2% | 54.8% | 48.0% | 52.0% | 47.2% | 52.8% | 50.4% | 49.6% |
Reproduce
All the code can be found in the ./reproduce directory.
Step-0 Extract Unique Contexts
First, we need to extract unique contexts in the datasets.
Code
```python def extract_unique_contexts(input_directory, output_directory): os.makedirs(output_directory, exist_ok=True) jsonl_files = glob.glob(os.path.join(input_directory, '*.jsonl')) print(f"Found {len(jsonl_files)} JSONL files.") for file_path in jsonl_files: filename = os.path.basename(file_path) name, ext = os.path.splitext(filename) output_filename = f"{name}_unique_contexts.json" output_path = os.path.join(output_directory, output_filename) unique_contexts_dict = {} print(f"Processing file: {filename}") try: with open(file_path, 'r', encoding='utf-8') as infile: for line_number, line in enumerate(infile, start=1): line = line.strip() if not line: continue try: json_obj = json.loads(line) context = json_obj.get('context') if context and context not in unique_contexts_dict: unique_contexts_dict[context] = None except json.JSONDecodeError as e: print(f"JSON decoding error in file {filename} at line {line_number}: {e}") except FileNotFoundError: print(f"File not found: {filename}") continue except Exception as e: print(f"An error occurred while processing file {filename}: {e}") continue unique_contexts_list = list(unique_contexts_dict.keys()) print(f"There are {len(unique_contexts_list)} unique `context` entries in the file {filename}.") try: with open(output_path, 'w', encoding='utf-8') as outfile: json.dump(unique_contexts_list, outfile, ensure_ascii=False, indent=4) print(f"Unique `context` entries have been saved to: {output_filename}") except Exception as e: print(f"An error occurred while saving to the file {output_filename}: {e}") print("All files have been processed.") ```Step-1 Insert Contexts
For the extracted contexts, we insert them into the LightRAG system.
Code
```python def insert_text(rag, file_path): with open(file_path, mode='r') as f: unique_contexts = json.load(f) retries = 0 max_retries = 3 while retries < max_retries: try: rag.insert(unique_contexts) break except Exception as e: retries += 1 print(f"Insertion failed, retrying ({retries}/{max_retries}), error: {e}") time.sleep(10) if retries == max_retries: print("Insertion failed after exceeding the maximum number of retries") ```Step-2 Generate Queries
We extract tokens from the first and the second half of each context in the dataset, then combine them as dataset descriptions to generate queries.
Code
```python tokenizer = GPT2Tokenizer.from_pretrained('gpt2') def get_summary(context, tot_tokens=2000): tokens = tokenizer.tokenize(context) half_tokens = tot_tokens // 2 start_tokens = tokens[1000:1000 + half_tokens] end_tokens = tokens[-(1000 + half_tokens):1000] summary_tokens = start_tokens + end_tokens summary = tokenizer.convert_tokens_to_string(summary_tokens) return summary ```Step-3 Query
For the queries generated in Step-2, we will extract them and query LightRAG.
Code
```python def extract_queries(file_path): with open(file_path, 'r') as f: data = f.read() data = data.replace('**', '') queries = re.findall(r'- Question \d+: (.+)', data) return queries ```Code Structure
.
├── examples
│ ├── batch_eval.py
│ ├── generate_query.py
│ ├── graph_visual_with_html.py
│ ├── graph_visual_with_neo4j.py
│ ├── lightrag_api_openai_compatible_demo.py
│ ├── lightrag_azure_openai_demo.py
│ ├── lightrag_bedrock_demo.py
│ ├── lightrag_hf_demo.py
│ ├── lightrag_lmdeploy_demo.py
│ ├── lightrag_ollama_demo.py
│ ├── lightrag_openai_compatible_demo.py
│ ├── lightrag_openai_demo.py
│ ├── lightrag_siliconcloud_demo.py
│ └── vram_management_demo.py
├── lightrag
│ ├── kg
│ │ ├── __init__.py
│ │ └── neo4j_impl.py
│ ├── __init__.py
│ ├── base.py
│ ├── lightrag.py
│ ├── llm.py
│ ├── operate.py
│ ├── prompt.py
│ ├── storage.py
│ └── utils.py
├── reproduce
│ ├── Step_0.py
│ ├── Step_1_openai_compatible.py
│ ├── Step_1.py
│ ├── Step_2.py
│ ├── Step_3_openai_compatible.py
│ └── Step_3.py
├── .gitignore
├── .pre-commit-config.yaml
├── Dockerfile
├── get_all_edges_nx.py
├── LICENSE
├── README.md
├── requirements.txt
├── setup.py
├── test_neo4j.py
└── test.pyStar History
Contribution
Thank you to all our contributors!
🌟Citation
@article{guo2024lightrag,
title={LightRAG: Simple and Fast Retrieval-Augmented Generation},
author={Zirui Guo and Lianghao Xia and Yanhua Yu and Tu Ao and Chao Huang},
year={2024},
eprint={2410.05779},
archivePrefix={arXiv},
primaryClass={cs.IR}
}Thank you for your interest in our work!








