CEdecisiontree 
An R package for lightweight cost-effectiveness analysis using decision trees.
Requests and comments welcome; please use Issues.
Installing CEdecisiontree
To install the development version from github:
library(remotes)
install_github("Health-Economics-in-R/CEdecisiontree")Then, to load the package, use:
library(CEdecisiontree)Motivation
Decisions trees can be modelled as special cases of more general models using available packages in R e.g. heemod, mstate or msm. Further, full probability models could be fit using a Bayesian model with e.g. Stan, jags or WinBUGS. However, simple decision tree models are often built in Excel, using statistics from literature or expert knowledge. This package is a analogue to these, such that models can be specified in a very similar and simple way.
Calculation
A decision tree is defined by parent-child pairs, i.e. from-to
connections, and the probability and associated value (e.g. cost) of
traversing each of the connections. Denote the probability of
transitioning from node
to
as
and the cost attributable to node
as
.
Where no connection exists between two nodes we shall say that the
parent’s set of children is the empty set
.
Denote the set of children by
.
Clearly, there are no
or
in this case but for computational purposes we will assume that
and
.
The expected value at each node
is calculated by ‘folding back’ using the recursive formula
with boundary values at the terminal nodes
Basic example
Quietly load libraries.
library(CEdecisiontree)
library(readr)
library(dplyr)
library(reshape2)
library(tidyr)
library(assertthat)We will consider a simple 7 node binary tree. Numeric labels are shown above each node. Probabilities and costs are show above and below each branch, respectively.
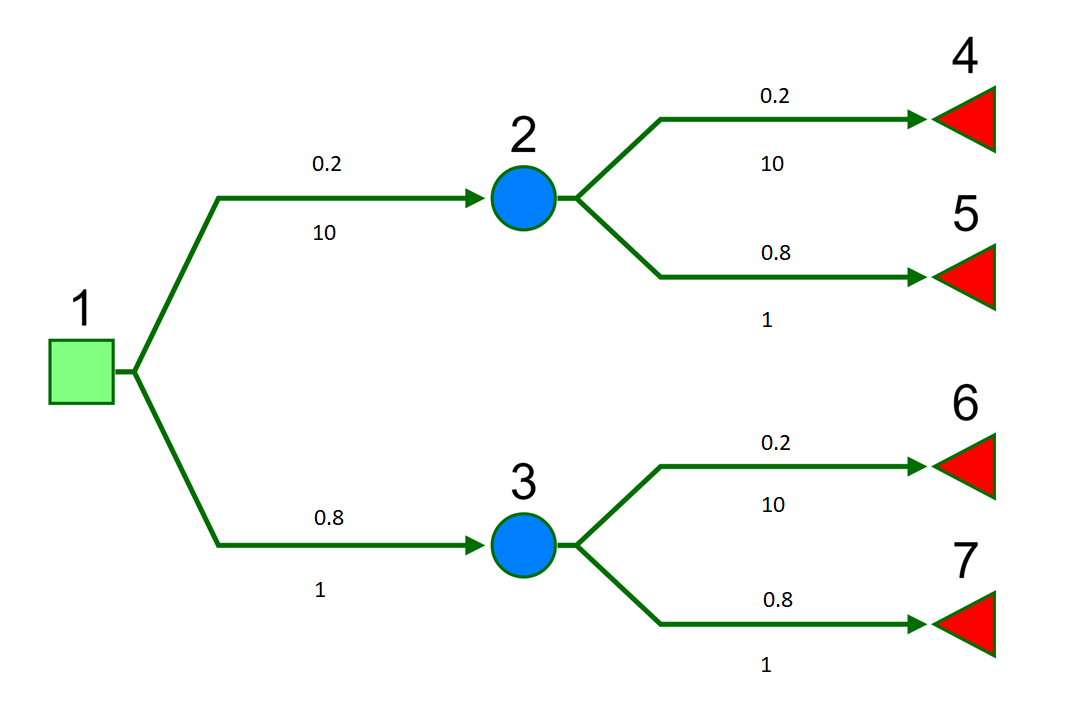
So if we were to write out the expected cost in full this would give
Load example data from the package.
data("cost")
data("probs")The cost and probability matrices we will use in this example are sparse
arrays indicating the edge values (rows=from node, columns=to node).
There are therefore the same dimensions and have the same entry pattern.
Empty cells have NA.
cost
#> # A tibble: 3 × 7
#> `1` `2` `3` `4` `5` `6` `7`
#> <dbl> <int> <int> <int> <int> <int> <int>
#> 1 NA 10 1 NA NA NA NA
#> 2 NA NA NA 10 1 NA NA
#> 3 NA NA NA NA NA 10 1probs
#> # A tibble: 3 × 7
#> `1` `2` `3` `4` `5` `6` `7`
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 NA 0.2 0.8 NA NA NA NA
#> 2 NA NA NA 0.2 0.8 NA NA
#> 3 NA NA NA NA NA 0.2 0.8probs is a probability transition matrix. This is like pmatrix.msm
in the msm package, or define_transition in the heemod package.
The transMat() function in the mstate package creates a closely
related multi-state model transition matrix. Copying this package, we
can create a decision tree transition matrix to use with this.
CEdecisiontree:::trans_binarytree(depth = 3)
#> to
#> from 1 2 3 4 5 6 7
#> 1 NA 1 2 NA NA NA NA
#> 2 NA NA NA 3 4 NA NA
#> 3 NA NA NA NA NA 5 6The expected value at each node is calculate as follows.
my_model <-
define_model(
transmat = list(vals = cost,
prob = probs))
dectree_expected_values(model = my_model)
#> 1 2 3 4 5 6 7
#> 5.6 12.8 3.8 10.0 1.0 10.0 1.0There is also an Rcpp version of this function.
Cdectree_expected_values(vals = as.matrix(cost),
p = as.matrix(probs))Other tree statistics
For additional information, including for the purposes of model checking we can calculate other tree statistics. We can obtain the contributing cost as weighted by the chance of occurrence. This can be thought of as a trade-off between the raw, original cost and branch position.
wcost <- branch_joint_probs(my_model) * cost
wcost
#> 1 2 3 4 5 6 7
#> 1 NA 2 0.8 NA NA NA NA
#> 2 NA NA NA 0.4 0.16 NA NA
#> 3 NA NA NA NA NA 1.6 0.64We can check that this sums to the same total expected cost.
sum(wcost, na.rm = TRUE)
#> [1] 5.6We can also calculate the joint probabilities of traversing to each
terminal state using branch_joint_probs. This is useful when an
alternative model set-up is used such that total costs and health values
are assigned to these terminal nodes only. Here we assume node labelling
order from root such that terminal nodes are last.
n_from_nodes <- nrow(probs)
n_to_nodes <- ncol(probs)
terminal_states <- (n_from_nodes + 1):n_to_nodes
p_terminal_state <-
branch_joint_probs(my_model)[ ,terminal_states] %>%
colSums(na.rm = TRUE)
p_terminal_state
#> 4 5 6 7
#> 0.04 0.16 0.16 0.64
sum(p_terminal_state)
#> [1] 1See package vignettes for more details and examples.
Code of Conduct
Please note that the CEdecisiontree project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.
