U-Net for Brain Tumor Segmentation
A U-Net is a Convolutional Neural Network with an architecture consisting of a contracting path to capture context and a symmetric expanding path that enables precise localization. It is highly effective in segmentation. This U-Net model is developed for segmentation of Brain Tumor in MRI scans using Keras and MedPy using the BraTS dataset
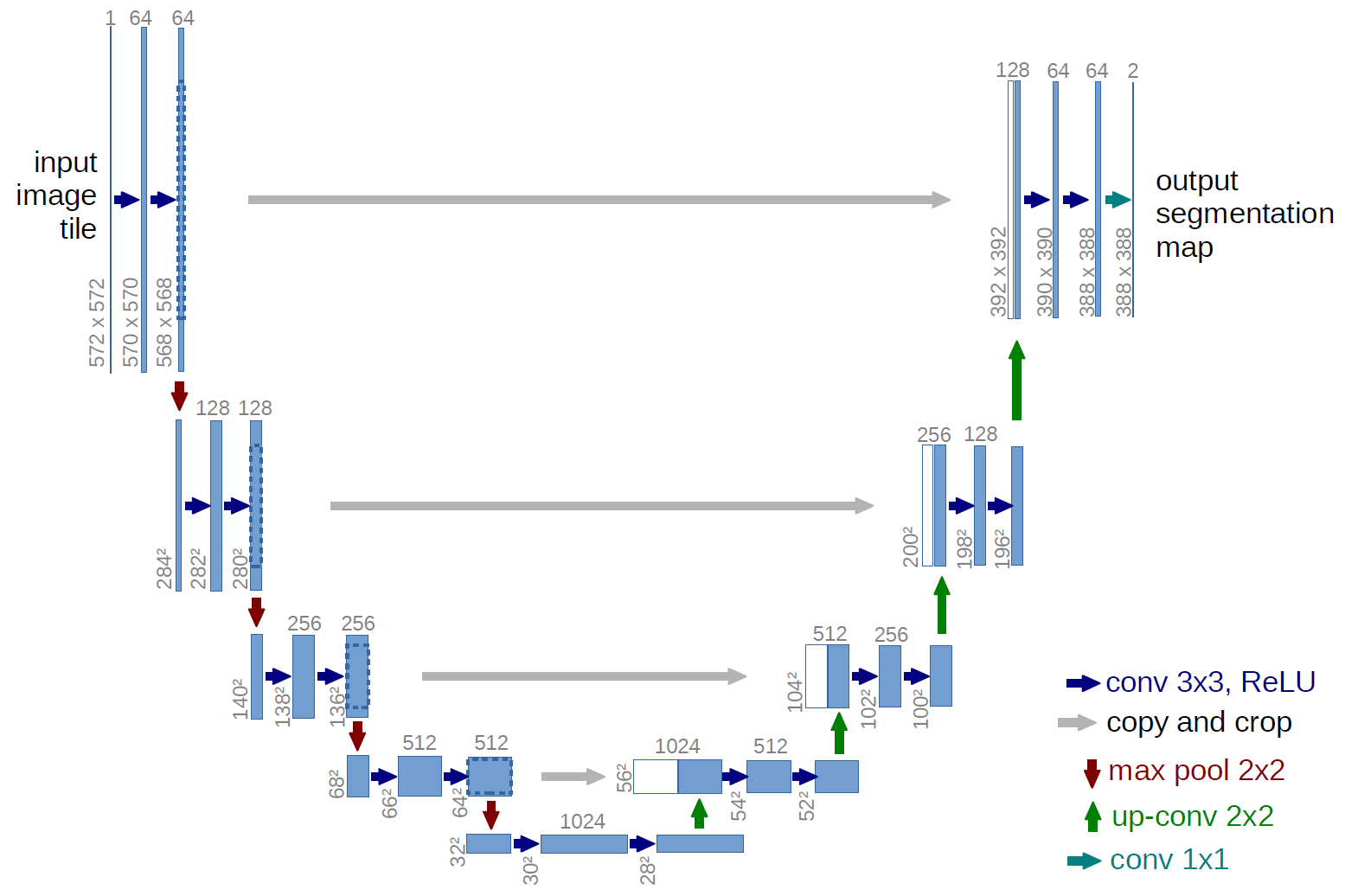
U-Net Architecture
Getting Started
Clone the repository and test the implementation using the fully trained model unet.hdf5
Prerequisites
The model requires the following modules to work
KerasMedpyThe implementation also requires access to BraTS dataset from MICCAI Challenge for training the model. Refer link below for more details
https://www.med.upenn.edu/sbia/brats2018/data.html
Installing
Follow the link below to install Keras libraries
https://keras.io/To install nytimesarticle API follow instruction in
https://pypi.org/project/MedPy/Running the tests
Pre-Processing data
MedPy package is installed in Python2 using scripts in (code-python2) and is used to load the data. The data is preprocessed and stored in a n256256*5 numpy array.
Change the API setting in the script as per the requirements
Training Model
Run code-python3/unet2D.py script to train the model.
NOTE: This steps requires Keras to be already installed
Authors
- Muthuvel Palanisamy - Initial work - muthu2093
See also the list of contributors who participated in this project.
License
This project is licensed under the MIT License - see the LICENSE.md file for details
References
[1] https://doi.org/10.7937/K9/TCIA.2017.KLXWJJ1Q