DIRECTORY
DaSSCo Mass Digitization App
Purpose of the application
The DaSSCo project is tasked with digitizing millions of specimens. To speed this process along, there needs to be a way to rapidly fill in data on 'storage', 'taxonomy', etc. The Mass Digitization App is here to achieve this goal.
Installation
Installation is done using a setup file that will ensure all dependencies are in place. The installer will also add a clean local database for registering entries in a "DaSSCo" folder under the user's documents folder. Be mindful to backup the database file upon reinstallation, so it is not overwritten and and data in it erased.
Downloads: (https://github.com/NHMDenmark/DaSSCo/releases/)
For Windows users: Please press the green tick mark icon in the process line under 'Show hidden icons' which represents 'Request administrator access'. Before installing you need to right-click the installation icon and select 'Run as administrator'. This will trigger a prompt to submit your KU credentials (Swedish license plate username + pass phrase).
Usage
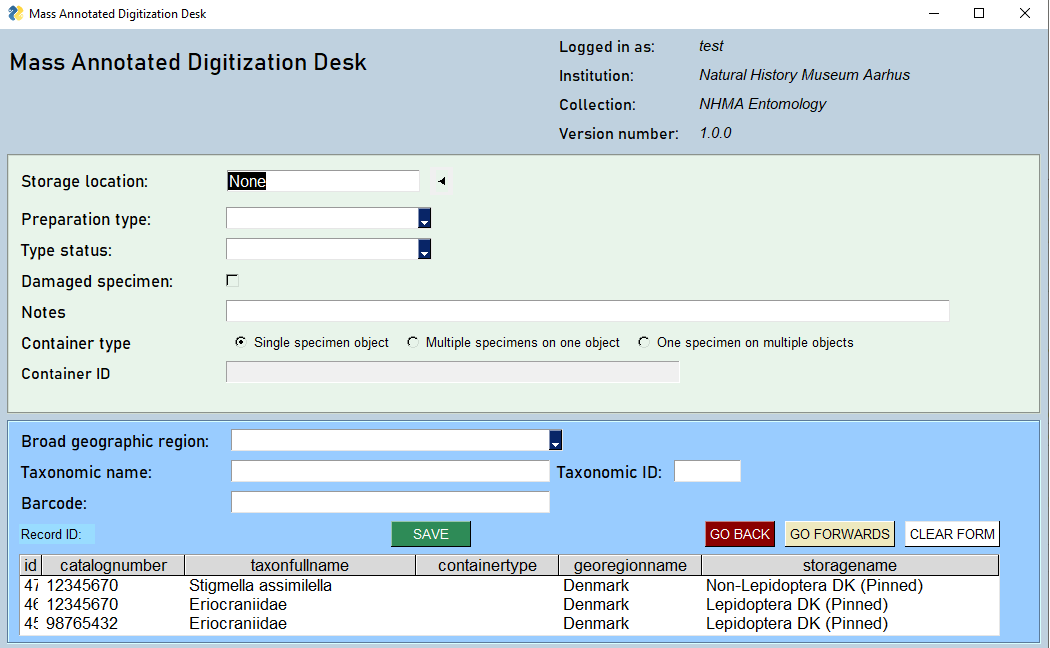
There is a path to follow that requires only little training. A user must have credentials in order to employ the app. One could say that a specimen record is being created bit by bit and submitted to local DB at the end.
Data entry
After log in, the first section focuses on specimen storage location which has an autosuggest feature. The path leads through prep type and status, and into Geographic region and taxonomy. The latter also has the auto suggest feature. Auto-suggest takes three keystroke to query among all the names and returns a row object of names. As the input to which the keystrokes contribute increases, the smaller the subset gets at which point it is feasible to arrow down through the result until the desired name is reached and press Enter.
The barcode is now ready for scanning. From there the record is ready to be 'saved'.
Novel taxon names
Should the case be that a taxon name is inputted which is not contained in the taxonomic spine - a pop-up window appears in which the novel name can be inputted. There follows a similar event where the higher taxonomic name is asked for. If a higher taxon name returns suggestions (must be family rank or higher) then arrow down to the desired name and press enter.
If the higher taxon name is also novel, then finish typing it and tab into the Cancel button. Press the spacebar to commit and the record will be registered with a taxonomic comment in the record notes field.
Alternative taxonomic identifiers
Certain collection will be using a different taxonomic name input system. For these specific cases a field "Taxonomic ID" will appear next to the main Taxonomic name input field. In this field you can add an identifier and press enter which populates the Taxonomic name field with the name corresponding to the identifier.
Specimen needs repair
A checkbox for damaged specimens has been addded to the app. Label = "Damaged specimen:". This should be checked if the specimen being digitized needs repair in any way.
Navigation between records
The "Back" and the "Forward" buttons are for paging through already entered records. You can follow the progress in the "Previous records table" near the bottom of the app. Once you go all the way back to the beginning of the records, you can press the Back-button again and the table will shoot to the top. The form will be populated by the latest (top) record. The app has a cyclical behavior in this regard.
Data export
As the app is designed to work "off line" all entries are stored locally. The entries thus registered can be exported to an Excel spreadsheet that can be imported into Specify. A smarter pipeline for getting the local data directly into Specify is currently being planned.
Licence and authorship
The application comes under the Apache-2.0 license which aligns with the Open Source and Open Science frameworks.
The authors of the application are :
Fedor A. Steeman, NHMD
Jan K. Legind, NHMD
Pip Brewer, NHMD
Systems Architecture
The app is written in python and consists of frontend components for easy user interfacing and backend component capable of both local storage and accessing external systems. The app is bundled with a local database file that is placed in a "DaSSCo" folder on the system it's installed on. In order to log on, and perform other functions, it needs to be connected to the internet. With the current state of the app, there are certain backend functions that have to be run by the developers in the development environment.
More information on the Systems Architecture including a visual representation, see here: Systems Architecture
For Developers
Structure
The app interfaces with a local SQLite database with tables for taxonomy (millions of names that are accessed according to the relevant discipline, say 'botany' for instance.) Storage while smaller also has its own table, as do Collection, Georegion and Institution. The table that is populated by the app is mainly 'specimen'. Eventually, the local DB instances will be uploaded to a server where the data will be processed into Specify. The application also interfaces directly with Specify through the Specify7 API (more information further below).
Specify Interface
In order to exchange information with Specify, the app has a module for interfacing with the Specify7 API. For now, this is mainly used for user authentication and basic info, but eventually this is planned to be built out into full synchronization in both directions.
Compilation
Remember to activate the python virtual environment (venv) and then first run:
pip install -r requirements.txtFor creating the executable, we use PyInstaller (https://pyinstaller.org/) using this command in the CLI from the root folder:
pyinstaller .\MassDigitizer\DaSSCo.py --onedir --noconsole --noconfirm --paths=MassDigitizer\For creating the installer, we use Inno Setup and a definition file is located in the repo root DaSSCO.iss. The Inno Setup scripts bundles the database with the executable into an installer file. Before running the Inno Setup script, it is necessary to fill the database file with the taxonomic spine and other predefined data.
Preparing Database File for Bundling
As part of the source code, there is a skeleton sqlite database file that needs to be filled with pre-defined data, not in the least the taxonomic spines for each collection. Before creating the installer, a temporary copy of the database file is generated that is then filled with the predefined data. Under MassDigitizer/sql/editions there are folders for each collection containing the SQL statements needed to insert not only taxonomic names, but also other predefined data specific for that collection such as storage locations, preparation types and type statuses.
This is done for two main reasons:
- To keep the size of the skeleton database on the repository below the 50MB limit
- To have version control of the predefined data by having it stored in SQL text files.
In the root of this folder there is a batch file (prepare-db.bat) that needs to be run in order to execute the sql statements. The resulting updated db.sqlite3 file is located in the temp folder from where it will be picked up by Inno Setup for being bundled with the installer.
In order to run the batch file, you need to install sqlite command tools first and make sure its path is added to your machine's PATH environment variables.
Collections currently supported
Here follows a table for the different collections supported so far:
| Institution | Collection | Path | Remarks |
|---|---|---|---|
| NHMD (Copenhagen) | Vascular Plants | NHMD\tracheophyta | |
| NHMD (Copenhagen) | Entomology | NHMD\entomology | Taxon spine restricted to selected taxa under Coleoptera & Lepidoptera |
| NHMA (Aarhus) | Entomology | NHMA\entomology |
Compilation process steps
So the process for compilation is as follows:
- Create the executable using PyInstaller
- Run the batch file to prepare the database
- Run the Inno Setup script to create the installer
The installer is then placed in folder Mass-Digitizer\MassDigitizer\Output ready for distribution. NOTE: For NHMD Vascular Plants, the taxon spine is recorded as sets of csv files that are placed in folder (Mass-Digitizer/tree/main/data/taxon spines/Botany). The respective SQL statements have been generated on the basis of those files using the python script (prepare-db.py). If changes are made to the taxon spine files, this script will need to be rerun. This will take many hours and should therefore not be a part of the standard compilation process.
Author backfill
Adding authors to Digi app exports that were made pre 1.1.3
The module author_backfill_with_test.py can be run with only two changes. The path to the SQLite DB must be set, as well as the xlsxwriter.Workbook name.
con = sqlite3.connect(r'C:\DaSSCo\db.sqlite3') #Please set the connection to the operational database (SQLite assumed).
workbook = xlsxwriter.Workbook("authorDropdown02.xlsx") # Output Excel sheet with the parameter name.Monitor script
Detecting new files coming into the 2.PostProcessed_openRefine directory and adding the three new columns described here: #461
The structure of the new columns is this:
- datafile_remark (the filename)
- datafile_source (a description of the raison d'ëtre of the remark, e.g. "DaSSCo datafile" or some such)
- datafile_date (I suppose this should be the date of export of the datafile)