
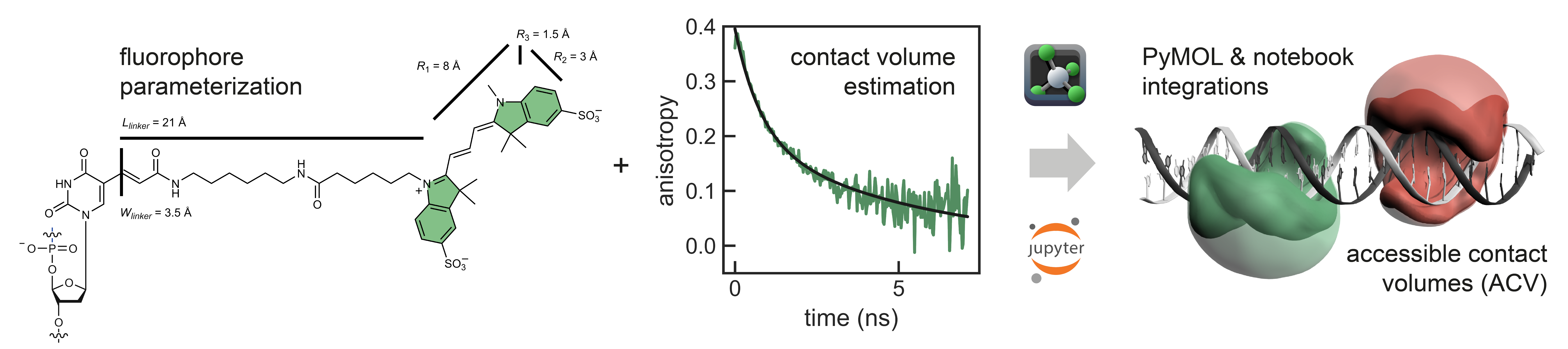
FRETraj is a Python module for predicting FRET efficiencies by calculating multiple accessible-contact volumes (multi-ACV) to estimate donor and acceptor dye dynamics. The package features a user-friendly PyMOL plugin1 for for FRET-assisted, integrative structural modeling. It interfaces with the LabelLib library for fast computation of ACVs. Specifically, FRETraj is designed to:
- plan (single-molecule) FRET experiments by optimizing labeling positions
- interpret FRET-based distance measurements on a biomolecule
- integrate FRET experiments with molecular dynamics simulations
Installation and Documentation
Follow the instructions for your platform here
References
If you use FRETraj in your work please refer to the following paper:
Additional readings
- F.D. Steffen, R.K.O. Sigel, R. Börner, Phys. Chem. Chem. Phys. 2016, 18, 29045-29055.
- S. Kalinin, T. Peulen, C.A.M. Seidel et al. Nat. Methods, 2012, 9, 1218-1225.
- T. Eilert, M. Beckers, F. Drechsler, J. Michaelis, Comput. Phys. Commun., 2017, 219, 377–389.
- M. Dimura, T. Peulen, C.A.M. Seidel et al. Curr. Opin. Struct. Biol. 2016, 40, 163-185.
- M. Dimura, T. Peulen, C.A.M Seidel et al. Nat. Commun. 2020, 11, 5394.
1 PyMOL was developed by WarrenDeLano and is maintained by Schrödinger, LLC.



