This package implements fast/scalable node embedding algorithms. This can be used to embed the nodes in graph objects and arbitrary scipy CSR Sparse Matrices. We support NetworkX graph types natively.
Installing
pip install nodevectors
This package depends on the CSRGraphs package, which is automatically installed along it using pip. Most development happens there, so running pip install --upgrade csrgraph once in a while can update the underlying graph library.
Supported Algorithms
-
Node2Vec (paper). Note that despite popularity this isn't always the best method. We recommend trying ProNE or GGVec if you run into issues.
-
GGVec (paper upcoming). A flexible default algorithm. Best on large graphs and for visualization.
-
ProNE (paper). The fastest and most reliable sparse matrix/graph embedding algorithm.
-
GLoVe (paper). This is useful to embed sparse matrices of positive counts, like word co-occurence.
-
Any Scikit-Learn API model that supports the
fit_transformmethod with then_componentattribute (eg. all manifold learning models, UMAP, etc.). Used with theSKLearnEmbedderobject.
Quick Example:
import networkx as nx
from nodevectors import Node2Vec
# Test Graph
G = nx.generators.classic.wheel_graph(100)
# Fit embedding model to graph
g2v = Node2Vec(
n_components=32,
walklen=10
)
# way faster than other node2vec implementations
# Graph edge weights are handled automatically
g2v.fit(G)
# query embeddings for node 42
g2v.predict(42)
# Save and load whole node2vec model
# Uses a smart pickling method to avoid serialization errors
# Don't put a file extension after the `.save()` filename, `.zip` is automatically added
g2v.save('node2vec')
# You however need to specify the extension when reading it back
g2v = Node2Vec.load('node2vec.zip')
# Save model to gensim.KeyedVector format
g2v.save_vectors("wheel_model.bin")
# load in gensim
from gensim.models import KeyedVectors
model = KeyedVectors.load_word2vec_format("wheel_model.bin")
model[str(43)] # need to make nodeID a str for gensim
Warning: Saving in Gensim format is only supported for the Node2Vec model at this point. Other models build a Dict or embeddings.
Embedding a large graph
NetworkX doesn't support large graphs (>500,000 nodes) because it uses lots of memory for each node. We recommend using CSRGraphs (which is installed with this package) to load the graph in memory:
import csrgraph as cg
import nodevectors
G = cg.read_edgelist("path_to_file.csv", directed=False, sep=',')
ggvec_model = nodevectors.GGVec()
embeddings = ggvec_model.fit_transform(G)The read_edgelist can take all the file-reading parameters of pandas.read_csv. You can also specify whether the graph is undirected (so all edges go both ways) or directed (so edges are one-way)
Best algorithms to embed a large graph
The ProNE and GGVec algorithms are the fastest. GGVec uses the least RAM to embed larger graphs. Additionally here are some recommendations:
-
Don't use the
return_weightandneighbor_weightif you are using the Node2Vec algorithm. It necessarily makes the walk generation step 40x-100x slower. -
If you are using GGVec, keep
orderat 1. Using higher order embeddings will take quadratically more time. Additionally, keepnegative_ratiolow (~0.05-0.1),learning_ratehigh (~0.1), and use aggressive early stopping values. GGVec generally only needs a few (less than 100) epochs to get most of the embedding quality you need. -
If you are using ProNE, keep the
stepparameter low. -
If you are using GraRep, keep the default embedder (TruncatedSVD) and keep the order low (1 or 2 at most).
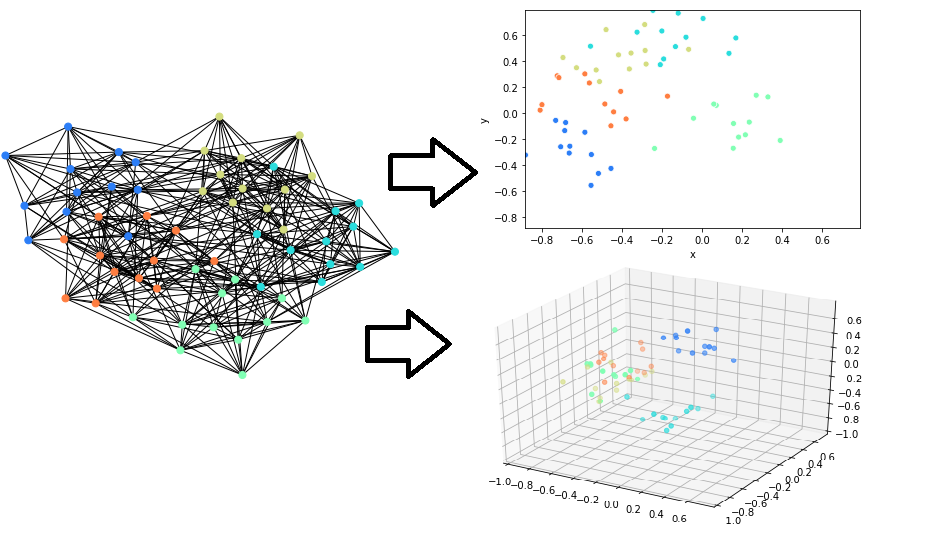
Preprocessing to visualize large graphs
You can use our algorithms to preprocess data for algorithms like UMAP or T-SNE. You can first embed the graph to 16-400 dimensions then use these embeddings in the final visualization algorithm.
Here is an example of this on the full english Wikipedia link graph (6M nodes) by Owen Cornec:

The GGVec algorithm often produces the best visualizations, but can have some numerical instability with very high n_components or too high negative_ratio. Node2Vec tends to produce elongated and filamented structures in the visualizations due to the embedding graph being sampled on random walks.
Embedding a VERY LARGE graph
(Upcoming).
GGVec can be used to learn embeddings directly from an edgelist file (or stream) when the order parameter is constrained to be 1. This means you can embed arbitrarily large graphs without ever loading them entirely into RAM.
Related Projects
-
DGL for Graph Neural networks.
-
KarateClub for node embeddings specifically on NetworkX graphs. The implementations are less scalable, because of it, but the package has more types of embedding algorithms.
-
GraphVite is not a python package but has GPU-enabled embedding algorithm implementations.
-
Cleora, another fast/scalable node embedding algorithm implementation
Why is it so fast?
We leverage CSRGraphs for most algorithms. This uses CSR graph representations and a lot of Numba JIT'ed procedures.