PIMENTA
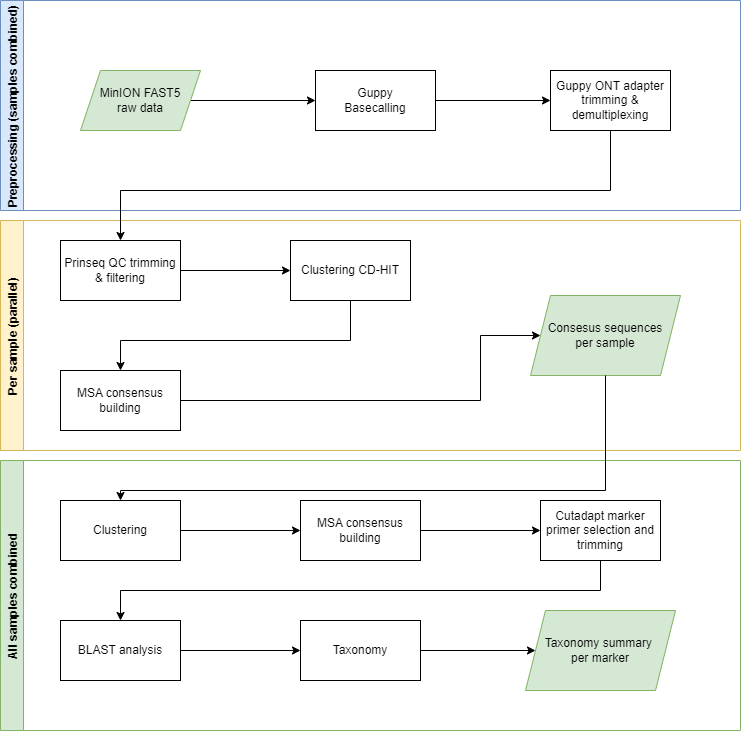
PIMENTA is a pipeline for the rapid identification of species in forensic samples using MinION data. The pipeline consists of eight linked tools, and data analysis passes through 3 phases: 1) pre-processing the MinION data through read calling, demultiplexing, trimming sequencing adapters, quality trimming and filtering the reads, 2) clustering the reads, continued by MSA and consensus building per cluster, 3) reclustering of consensus sequences, followed by another MSA and consensus building per cluster, 4) Taxonomy identification with the use of a BLAST analysis. PIMENTA makes use of the frequently-used software tools Cutadapt v4.2 (http://cutadapt.readthedocs.io/en/stable/guide.html), PRINSEQ v0.20.4 (http://prinseq.sourceforge.net/), BLAST+ (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/), Guppy 6.5,7, CD-HIT v4.8.1 (https://github.com/weizhongli/cdhit), Consensus v1.0, Krona v2.7.1 (https://github.com/marbl/Krona/wiki) and MAFFT v7.130 (https://mafft.cbrc.jp/alignment/software/).
If you use PIMENTA, please cite: https://doi.org/10.1101/2024.02.14.580249
Required programs
Mininconda and has to be installed and loaded before installing PIMENTA.
Guppy == 6.5.7
Installing the pipeline
First clone the pipeline into a chosen directory:
git clone https://github.com/WFSRDataScience/PIMENTA.gitTo install PIMENTA:
cd PIMENTA
bash install.sh Required R packages
dplyr, readr, stringi, taxonomizr, stringr and data.table are required for Taxonomic_summary.R
The required R packages are automatically installed.
Required perl packages
Bio::SeqIO, Bio::AlignIO
These are automatically installed with install.sh
Required databases
Taxdump database, download and extract the files in a desired location: https://ftp.ncbi.nlm.nih.gov/pub/taxonomy/new_taxdump/
The variable NT_dmp in the settingsfile is where you specify where you extracted the dmp files to.
NCBI NT database, specify the location in DATABASE variable in settingsfile.
update_blastdb.pl --decompress ntGeneral usage The basic command to run the example analysis, after editing the settings in settingsfile.txt:
bash START.sh settingsfile.txtThe previous command runs Guppy and the first clustering per sample. To run the reclustering and Taxonomic identification:
bash START_recluster.sh settingsfile.txtModify the following parameters in settingsfile.txt to run the pipeline:
<RunName> Run name of the analysis, this will be the name of the output folder
<mail-user> Email address to receive updates about the job and results from the analysis
<modus> "all" to analyse all datasets within OutDir or "one" to analyse dataset with same RunName and OutDir
<OutDir> Path where the output of the DNA metabarcoding analysis is placed
<workdir> Location of the PIMENTA scripts on the HPC server
<THREADS> Total CPU threads
Other options:
<RunModules> Modules to run, options: all (default), Guppy,Clustering, Consensus, Blast, Taxonomy.
Another module called 'oldmode' can be also be used, which runs the tax identification per sample (instead of per dataset). Needs to be used in combination with 'all' or other modules (e.g. "oldmode,clustering,Consensus,Blast,taxonomy") (experimental)
<GPU> GPU usage (only for Guppy basecalling): '1' for using GPU or 'cpu' to run Guppy on CPU.
Output files
By default, the output of PIMENTA is written to /<OutDir>/<RunName>
The taxonomy and QC output is written to /<OutDir>/<RunName>/Taxonomy_persample<ident2>
Disclaimer
Although the authors of this pipeline have taken care to consider exceptions such as incorrectly annotated sequence records in public databases, taxonomic synonyms, and ambiguities, the user is advised that the results of this pipeline can in no way be construed as conclusive evidence for either positive or negative taxonomic identification of the contents of biological materials. The pipeline and the results it produces are provided for informational purposes only. To emphasize this point, we reproduce the disclaimer of the license under which this pipeline is released verbatim, below:
THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS" AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT HOLDER OR CONTRIBUTORS BE LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.