A library for data valuation.
pyDVL collects algorithms for Data Valuation and Influence Function computation. Here is the list of all methods implemented.
Data Valuation for machine learning is the task of assigning a scalar to each element of a training set which reflects its contribution to the final performance or outcome of some model trained on it. Some concepts of value depend on a specific model of interest, while others are model-agnostic. pyDVL focuses on model-dependent methods.

Comparison of different data valuation methods on best sample removal.
The Influence Function is an infinitesimal measure of the effect that single training points have over the parameters of a model, or any function thereof. In particular, in machine learning they are also used to compute the effect of training samples over individual test points.
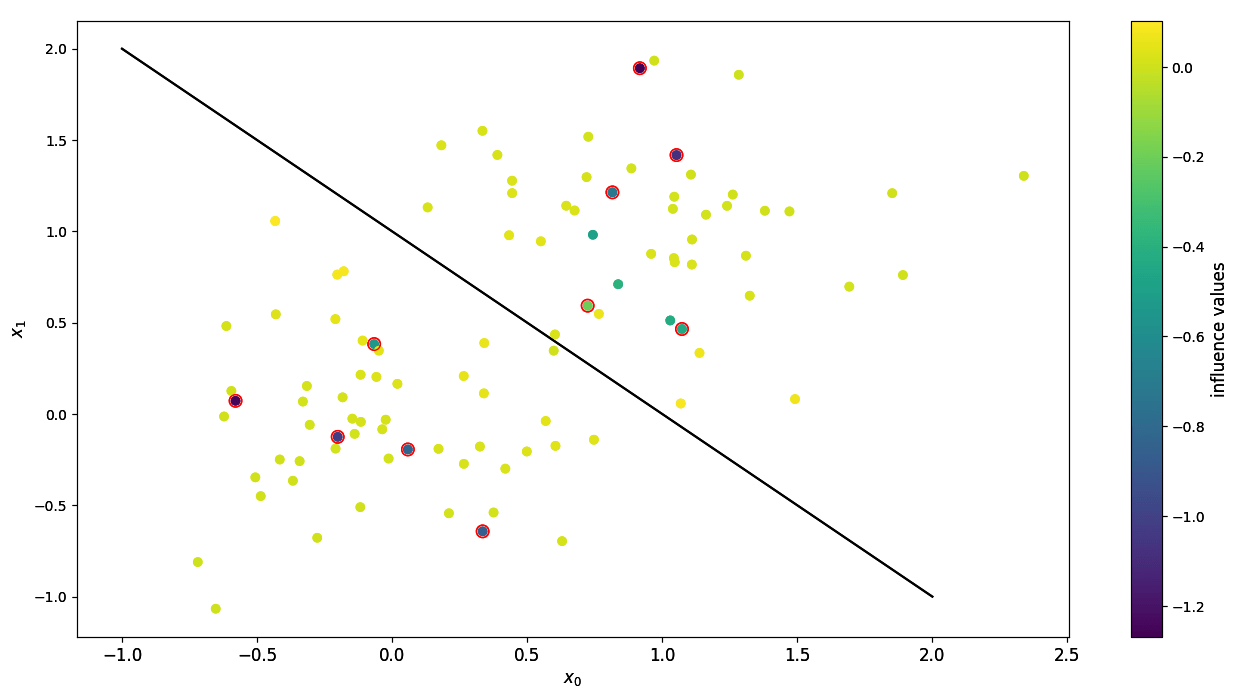
Influences of input points with corrupted data. Highlighted points have flipped labels.
Installation
To install the latest release use:
$ pip install pyDVLYou can also install the latest development version from TestPyPI:
pip install pyDVL --index-url https://test.pypi.org/simple/pyDVL has also extra dependencies for certain functionalities, e.g. for using influence functions run
$ pip install pyDVL[influence]For more instructions and information refer to Installing pyDVL in the documentation.
Usage
Please read Getting Started in the documentation for more instructions. We provide several examples for data valuation and for influence functions in our Example Gallery.
Influence Functions
- Import the necessary packages (the exact ones depend on your specific use case).
- Create PyTorch data loaders for your train and test splits.
- Instantiate your neural network model and define your loss function.
- Instantiate an
InfluenceFunctionModeland fit it to the training data - For small input data, you can call the
influences()method on the fitted instance. The result is a tensor of shape(training samples, test samples)that contains at index(i, j) the influence of training sampleion test samplej. - For larger datasets, wrap the model into a "calculator" and call methods on it. This splits the computation into smaller chunks and allows for lazy evaluation and out-of-core computation.
The higher the absolute value of the influence of a training sample on a test sample, the more influential it is for the chosen test sample, model and data loaders. The sign of the influence determines whether it is useful (positive) or harmful (negative).
Note pyDVL currently only support PyTorch for Influence Functions. We plan to add support for Jax next.
import torch
from torch import nn
from torch.utils.data import DataLoader, TensorDataset
from pydvl.influence import SequentialInfluenceCalculator
from pydvl.influence.torch import DirectInfluence
from pydvl.influence.torch.util import (
NestedTorchCatAggregator,
TorchNumpyConverter,
)
input_dim = (5, 5, 5)
output_dim = 3
train_x, train_y = torch.rand((10, *input_dim)), torch.rand((10, output_dim))
test_x, test_y = torch.rand((5, *input_dim)), torch.rand((5, output_dim))
train_data_loader = DataLoader(TensorDataset(train_x, train_y), batch_size=2)
test_data_loader = DataLoader(TensorDataset(test_x, test_y), batch_size=1)
model = nn.Sequential(
nn.Conv2d(in_channels=5, out_channels=3, kernel_size=3),
nn.Flatten(),
nn.Linear(27, 3),
)
loss = nn.MSELoss()
infl_model = DirectInfluence(model, loss, hessian_regularization=0.01)
infl_model = infl_model.fit(train_data_loader)
# For small datasets, instantiate the full influence matrix:
influences = infl_model.influences(test_x, test_y, train_x, train_y)
# For larger datasets, use the Influence calculators:
infl_calc = SequentialInfluenceCalculator(infl_model)
# Lazy object providing arrays batch-wise in a sequential manner
lazy_influences = infl_calc.influences(test_data_loader, train_data_loader)
# Trigger computation and pull results to memory
influences = lazy_influences.compute(aggregator=NestedTorchCatAggregator())
# Trigger computation and write results batch-wise to disk
lazy_influences.to_zarr("influences_result", TorchNumpyConverter())Data Valuation
The steps required to compute data values for your samples are:
- Import the necessary packages (the exact ones will depend on your specific use case).
- Create a
Datasetobject with your train and test splits. - Create an instance of a
SupervisedModel(basically any sklearn compatible predictor), and wrap it in aUtilityobject together with the data and a scoring function. - Use one of the methods defined in the library to compute the values. In the
example below, we will use Permutation Montecarlo Shapley, an approximate
method for computing Data Shapley values. The result is a variable of type
ValuationResultthat contains the indices and their values as well as other attributes. - Convert the valuation result to a dataframe, and analyze and visualize the values.
The higher the value for an index, the more important it is for the chosen model, dataset and scorer. Reciprocally, low-value points could be mislabelled, or out-of-distribution, and dropping them can improve the model's performance.
from sklearn.datasets import load_breast_cancer
from sklearn.linear_model import LogisticRegression
from pydvl.utils import Dataset, Scorer, Utility
from pydvl.value import (MaxUpdates, RelativeTruncation,
permutation_montecarlo_shapley)
data = Dataset.from_sklearn(
load_breast_cancer(),
train_size=10,
stratify_by_target=True,
random_state=16,
)
model = LogisticRegression()
u = Utility(
model,
data,
Scorer("accuracy", default=0.0)
)
values = permutation_montecarlo_shapley(
u,
truncation=RelativeTruncation(u, 0.05),
done=MaxUpdates(1000),
seed=16,
progress=True
)
df = values.to_dataframe(column="data_value")Contributing
Please open new issues for bugs, feature requests and extensions. You can read about the structure of the project, the toolchain and workflow in the guide for contributions.
License
pyDVL is distributed under LGPL-3.0. A complete version can be found in two files: here and here.
All contributions will be distributed under this license.



