1. DeepEventMine
A deep leanring model to predict named entities, triggers, and nested events from biomedical texts.
- The model and results are reported in our paper:
DeepEventMine: End-to-end Neural Nested Event Extraction from Biomedical Texts, Bioinformatics, 2020.
1.1. Features
- Based on pre-trained BERT
- Predict nested entities and nested events
- Provide our trained models on the seven biomedical tasks
- Reproduce the results reported in our Bioinformatics paper
- Predict for new data given raw text input or PubMed ID
- Visualize the predicted entities and events on the brat
1.2. Tasks
- DeepEventMine has been trained and evaluated on the following tasks (six BioNLP shared tasks and MLEE).
- cg: Cancer Genetics (CG), 2013
- ge11: GENIA Event Extraction (GENIA), 2011
- ge13: GENIA Event Extraction (GENIA), 2013
- id: Infectious Diseases (ID), 2011
- epi: Epigenetics and Post-translational Modifications (EPI), 2011
- pc: Pathway Curation (PC), 2013
- mlee: Multi-Level Event Extraction (MLEE)
1.3. Our trained models and scores
2. Preparation
2.1. Requirements
- Python 3.6.5
- PyTorch (torch==1.1.0 torchvision==0.3.0, cuda92)
virtualenv -p python3 pytorch-env
source pytorch-env/bin/activate
export CUDA_VISIBLE_DEVICES=0
CUDA_PATH=/usr/local/cuda pip install torch==1.1.0 torchvision==0.3.0- Install Python packages
sh install.sh2.2. BERT
- Download SciBERT BERT model from PyTorch AllenNLP
sh download.sh bert2.3. DeepEventMine
- Download pre-trained DeepEventMine model on a given task
- [task] = cg (or pc, ge11, epi, etc)
sh download.sh deepeventmine [task]2.4 Brat
sh download.sh brat- Install brat based on the brat instructions
cd brat/brat-v1.3_Crunchy_Frog/ ./install.sh -u python2 standalone.py
3. Predict (BioNLP tasks)
3.1. Prepare data
- Download corpora
- To download the original data sets from BioNLP shared tasks.
- [task] = cg, pc, ge11, etc
sh download.sh bionlp [task]-
Preprocess data
- Tokenize texts and prepare data for prediction
sh preprocess.sh bionlp
- Tokenize texts and prepare data for prediction
-
Generate configs
- If using GPU: [gpu] = 0, otherwise: [gpu] = -1
- [task] = cg, pc, etc
sh run.sh config [task] [gpu]
3.2. Predict
- For development and test sets (given gold entities)
- CG task: [task] = cg
- PC task: [task] = pc
- Similarly for: ge11, ge13, epi, id, mlee
sh run.sh predict [task] gold dev
sh run.sh predict [task] gold test- Check the output in the path
experiments/[task]/predict-gold-dev/ experiments/[task]/predict-gold-test/
3.3. Evaluate
-
Retrieve the original offsets and create zip format
sh run.sh offset [task] gold dev sh run.sh offset [task] gold test -
Submit the zipped file to the shared task evaluation sites:
- Evaluate events
- Evaluate event prediction for PC and CG tasks on the development sets using the shared task scripts.
- Evaluation options: s (softboundary), p(partialrecursive)
sh run.sh eval [task] gold dev sp4. End-to-end
4.1. Input: a single PMID or PMCID
-
Abstract
sh pubmed.sh e2e pmid 1370299 cg 0 -
Full text
sh pubmed.sh e2e pmcid PMC4353630 cg 0 -
Input: PMID: 1370299, PMCID: PMC4353630 (a single PubMed ID to get raw text)
-
Model to predict: DeepEventMine trained on cg (Cancer Genetics 2013), (other options: pc, ge11, etc)
-
GPU: 0 (if CPU: -1)
-
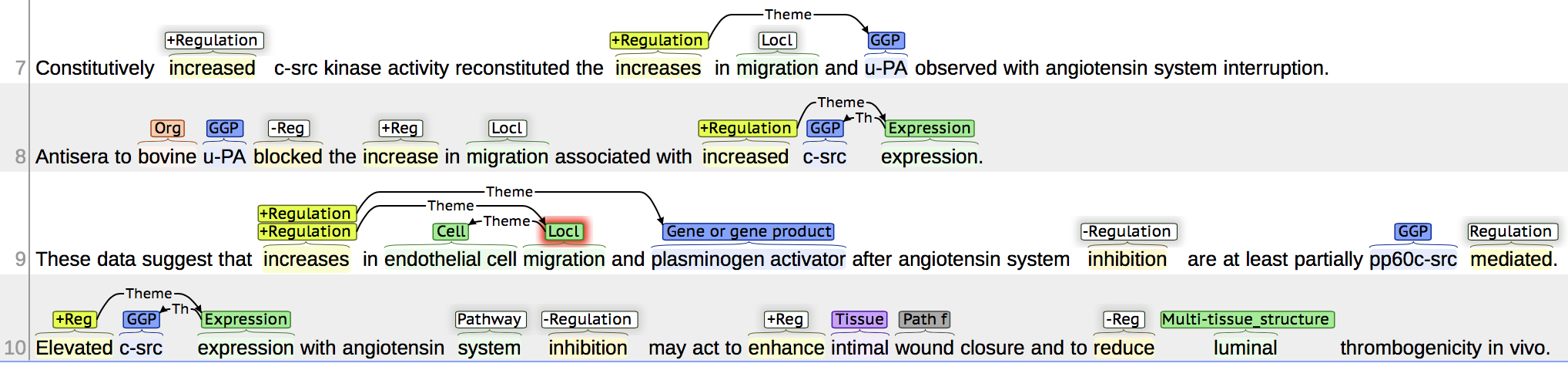
Output: in brat format and brat visualization
T24 Organism 1248 1254 bovine
T25 Gene_or_gene_product 1255 1259 u-PA
T55 Positive_regulation 1107 1116 increased
T57 Localization 1170 1179 migration
T58 Negative_regulation 1260 1267 blocked
...
T23 Gene_or_gene_product 1184 1188 u-PA
T56 Positive_regulation 1157 1166 increases
E9 Positive_regulation:T56 Theme:T23
T26 Gene_or_gene_product 1320 1325 c-src
T62 Gene_expression 1326 1336 expression
E10 Gene_expression:T62 Theme:T26
T61 Positive_regulation 1310 1319 increased
E24 Positive_regulation:T61 Theme:E10
## 4.2. Input: a list of PMIDs - Given an arbitrary name for your raw text data, for example "my-pubmed" - Prepare a list of PMID and PMCID in the path ```bash data/my-pubmed/pmid.txt ``` ```bash sh pubmed.sh e2e pmids my-pubmed cg 0 ``` ## 4.3. Input: raw text files - Given an arbitrary name for your raw text data, for example "my-pubmed" - Prepare your raw text files in the path ```bash data/my-pubmed/text/PMID-*.txt data/my-pubmed/text/PMC-*.txt ``` ```bash sh pubmed.sh e2e rawtext my-pubmed cg 0 ``` # 5. Predict for new data (step-by-step) - Input: your own raw text or PubMed ID - Output: predicted entities and events in brat format ## 5.1. Raw text - Given an arbitrary name for your raw text data, for example "my-pubmed" - Prepare your own raw text in the following path ```bash data/my-pubmed/text/PMID-*.txt data/my-pubmed/text/PMC-*.txt ``` ## 5.2. PubMed ID - Or, you can automatically get raw text given PubMed ID or PMC ID ### Get raw text 1. PubMed ID list - In order to get full text given PMC ID, the text should be available in ePub (for our current version). - Prepare your list of PubMed ID and PMC ID in the path ```bash data/my-pubmed/pmid.txt ``` - Get text from the PubMed ID ```bash sh pubmed.sh pmids my-pubmed ``` 2. PubMed ID - You can also get text by directly input a PubMed or PMC ID ```bash sh pubmed.sh pmid 1370299 sh pubmed.sh pmcid PMC4353630 ``` ### Preprocess ```bash sh pubmed.sh preprocess my-pubmed ``` ## 5.3. Predict 1. Generate config - Generate config for prediction - The data name to predict: my-pubmed - The trained model used for predict: cg (or pc, ge11, etc) - If you use gpu [gpu]=0, otherwise [gpu]=-1 ```bash sh pubmed.sh config my-pubmed cg 0 ``` 2. Predict ```bash sh pubmed.sh predict my-pubmed ``` 3. Retrieve the original offsets ```bash sh pubmed.sh offset my-pubmed ``` - Check the output in ```bash experiments/my-pubmed/results/ev-last/my-pubmed-brat ``` # 6. Visualization ## 6.1. Prepare data - Copy the predicted data into the brat folder to visualize - For the raw text prediction: ```bash sh pubmed.sh brat my-pubmed cg ``` - Or for the shared task ```bash sh run.sh brat [task] gold dev sh run.sh brat [task] gold test ``` ## 6.2. Visualize - The data to visualize is located in ```bash brat/brat-v1.3_Crunchy_Frog/data/my-pubmed-brat brat/brat-v1.3_Crunchy_Frog/data/[task]-brat ``` # 7. Acknowledgements This work is based on results obtained from a project commissioned by the New Energy and Industrial Technology Development Organization (NEDO). This work is also supported by PRISM (Public/Private R&D Investment Strategic Expansion PrograM). # 8. Citation ```bash @article{10.1093/bioinformatics/btaa540, author = {Trieu, Hai-Long and Tran, Thy Thy and Duong, Khoa N A and Nguyen, Anh and Miwa, Makoto and Ananiadou, Sophia}, title = "{DeepEventMine: End-to-end Neural Nested Event Extraction from Biomedical Texts}", journal = {Bioinformatics}, year = {2020}, month = {06}, issn = {1367-4803}, doi = {10.1093/bioinformatics/btaa540}, url = {https://doi.org/10.1093/bioinformatics/btaa540}, note = {btaa540}, eprint = {https://academic.oup.com/bioinformatics/article-pdf/doi/10.1093/bioinformatics/btaa540/33399046/btaa540.pdf}, } ```