ngs_te_mapper2: A program to identify transposable element insertions using next generation sequencing data
Table of Contents
Introduction
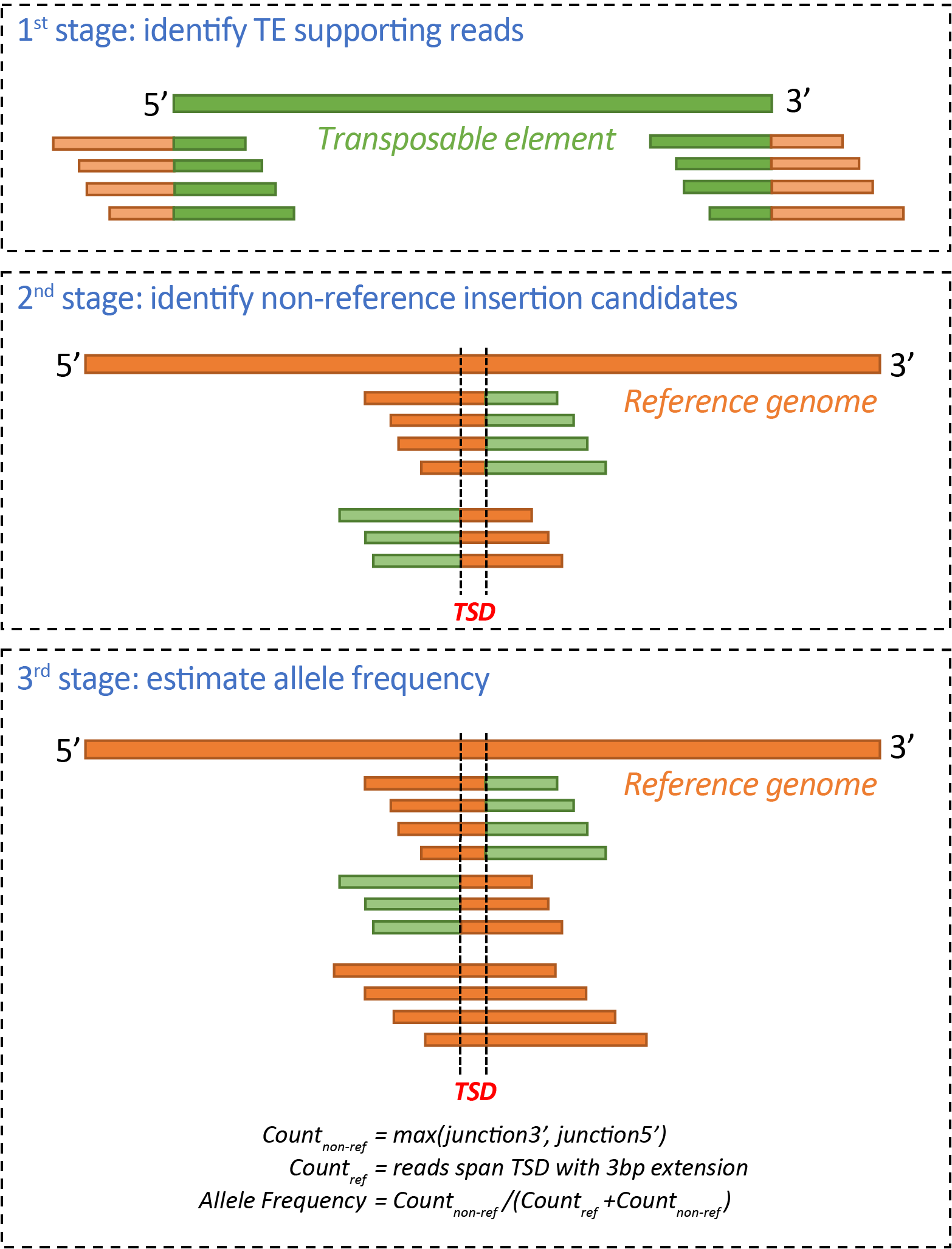
ngs_te_mapper2 is a method for detecting transposable element (TE) insertions from short-read next-generation sequencing (NGS) data described in Han et al. (2021) Genetics 219(2):iyab113. ngs_te_mapper2 is a Python re-implementation of the ngs_te_mapper method originally described in Linheiro and Bergman (2012) PLoS ONE 7(2): e30008. ngs_te_mapper2 uses a three-stage procedure to annotate non-reference TEs as the span of target site duplication (TSD), following the framework described in Bergman (2012) Mob Genet Elements. 2:51-54.
-
In the first stage, WGS reads are queried against a library of TE sequences to identify 'junction reads' that span the start/end of TE and genomic flanking sequences are retained. Such reads are often referred as 'split reads', although in reality these reads are not split in the resequenced genome.
-
In the second stage, junction reads from each side of TE insertion identified in the first stage are separately aligned to a reference genome that is hard-masked with RepeatMasker (http://www.repeatmasker.org/) using the same TE library from stage one. Genome-wide coverage profiles are computed using samtools v1.9 and genomic intervals with enriched coverage from junction read clusters on the 5' and 3' side of TEs are annotated in bed format. Regions of overlap between intervals of junction read clusters on the 5' and 3' side of TEs define the locations of the TSDs for candidate non-reference TE insertions. The orientation of the TE is determined from the relative orientation of alignments of the junction reads to the reference genome and TE library.
-
In the third stage, all reads from the original whole genome shotgun sequence data are used to query against the same hard-masked reference genome as in stage two. This additional mapping step is necessary to obtain all reads that span the TE-flank junction, as well as identify if any reads are present for the alternative ``reference" haplotype that does not carry the TE insertion. For each candidate non-reference TE insertion site, number of junction reads covering 5' and 3' side of each candidate TE insertion are estimated as the number of soft-clipped reads overlapping a 10bp window on the 5' and 3' side of the TSD, respectively (Count_junction5' and Count_junction3'). Number of non-reference reads (Count_non_ref) were estimated as max(Count_junction5', Count_junction3'). Number of reference reads (Count_ref) were estimated as number of non-soft-clipped reads spanning the TSD with at least 3bp extension on both side. The allele frequency for non-reference TEs is heuristically estimated as Count_non-ref/(Count_non_ref + Count_ref).
-
Reference TE insertions are detected using a similar strategy to non-reference insertions, independently of any reference TE annotation. The first stage in detecting reference TE insertions is identical to the first stage of detecting non-reference TE insertions described above. The second stage in identifying reference TE insertions involves alignment of the renamed, but otherwise unmodified, junction reads to the reference genome. Alignments of the complete junction read (i.e. non-TE and TE components) are clustered to identify the two ends of the reference TE insertion. The orientation of the reference TE is then determined from the relative orientation of alignments of the junction reads to the reference genome and TE library.
ngs_te_mapper2 is written in python3 and is designed to run on a Linux operating system.
Installation
Install Miniconda
To install ngs_te_mapper2, the recommended way is using conda. If your system doesn't have conda installed, you could use following steps to install Miniconda (Python 3.X). For more on Conda: see the Conda User Guide.
wget https://repo.continuum.io/miniconda/Miniconda3-latest-Linux-x86_64.sh -O $HOME//miniconda.sh
bash ~/miniconda.sh -b -p $HOME/miniconda # silent mode
echo "export PATH=\$PATH:\$HOME/miniconda/bin" >> $HOME/.bashrc # add to .bashrc
source $HOME/.bashrc
conda init # this step requires you to close and open a new terminal before it take effect
conda update conda # update condaInstall ngs_te_mapper2 using conda
ngs_te_mapper2 and all software dependencies can be installed using conda.
# We recommended installing ngs_te_mapper2 in a new conda environment
conda create -n ngs_te_mapper2 --channel bioconda ngs_te_mapper2
# Alternatively, you can install ngs_te_mapper2 in current active environment
conda install --channel bioconda ngs_te_mapper2Run ngs_te_mapper2 on test dataset
A test dataset is provided in the test/ directory, you can test whether your ngs_te_mapper2 installation is successful by running ngs_te_mapper2 on this dataset, which should take less than one minute to finish on a single thread machine.
conda activate ngs_te_mapper2
cd test
ngs_te_mapper2 -o test_output -f reads.fastq -r ref_1kb.fasta -l library.fastaNOTE: Sometimes activating conda environments does not work via conda activate myenv when run through a script submitted to a queueing system, this can be fixed by activating the environment in the script as shown below
CONDA_BASE=$(conda info --base)
source ${CONDA_BASE}/etc/profile.d/conda.sh
conda activate ngs_te_mapper2Usage
ngs_te_mapper2 required input files
- FASTQ File (
-f/--reads)- Raw reads from paired-end or single-end sequencing run in fastq or fastq.gz format.
- Multiple fastq/fastq.gz files can be provided separated by comma (ep.
-f R1.fasta,R2.fasta).
- TE library FASTA (
-l/--library)- A FASTA file containing a consensus sequence for each TE family. Note: Each family should only be represented in one sequence in this file.
- Example consensus FASTA file
- Reference FASTA (
-r/--reference)- The genome sequence of the reference genome in FASTA format.
Command line help page
usage: ngs_te_mapper2 [-h] -f READS -l LIBRARY -r REFERENCE [-a ANNOTATION]
[-n REGION] [-w WINDOW] [--min_mapq MIN_MAPQ]
[--min_af MIN_AF] [--tsd_max TSD_MAX]
[--gap_max GAP_MAX] [-m MAPPER] [-t THREAD] [-o OUT]
[-p PREFIX] [-k]
Script to detect non-reference TEs from single end short read data
required arguments:
-f READS, --reads READS
raw reads in fastq or fastq.gz format, separated by
comma
-l LIBRARY, --library LIBRARY
TE concensus sequence
-r REFERENCE, --reference REFERENCE
reference genome
optional arguments:
-h, --help show this help message and exit
-a ANNOTATION, --annotation ANNOTATION
reference TE annotation in GFF3 format (must have
'Target' attribute in the 9th column)
-w WINDOW, --window WINDOW
merge window for identifying TE clusters (default =
10)
--min_mapq MIN_MAPQ minimum mapping quality of alignment (default = 20)
--min_af MIN_AF minimum allele frequency (default = 0.1)
--tsd_max TSD_MAX maximum TSD size (default = 25)
--gap_max GAP_MAX maximum gap size (default = 5)
-t THREAD, --thread THREAD
thread (default = 1)
-o OUT, --out OUT output dir (default = '.')
-p PREFIX, --prefix PREFIX
output prefix
-k, --keep_files If provided then all intermediate files will be kept
(default: remove intermediate files)Note: The optional reference TE annotation input should in theory speed up the program. ngs_te_mapper2 expects the TE annotation to be in GFF3 format and Target attribute must be included in the 9th column that represents TE family name. If you have *.out annotation generated by RepeatMasker, you can use this utility script to convert from *.out to GFF3 format.
Output
ngs_te_mapper2 outputs reference and non-referece TE insertion predictions in BED format (0-based).
<sample>.nonref.bed: non-reference TE insertion annotation predicted by ngs_te_mapper2 pipeline in BED format (0-based).<sample>.ref.bed: reference TE insertion annotation predicted by ngs_te_mapper2 pipeline in BED format (0-based).
TE insertion annotation in bed format
ngs_te_mapper2 generates standard BED file <sample>.nonref.bed and <sample>.ref.bed that have detailed information for each reference and non-reference TE insertion.
| Column | Description |
|---|---|
| chromosome | The chromosome name where the TE insertion occurred |
| position | Starting breakpoint position of the TE insertions. |
| end | Ending breakpoint position of the TE insertions. |
| info | Includes TE family, TSD, Allele Frequency, 3' support, 5' support and reference reads. Separated by '|'. |
| score | '.' |
| strand | Strand that TE insertion occurs |
Log file output by ngs_te_mapper2
For each ngs_te_mapper2 run, a log file called <sample>.log is generated that records all the major steps in the program and error messages.
Getting help
Please use the Github Issue page if you have questions.
Citation
To cite ngs_te_mapper2 in publications, please use:
S. Han, P.J. Basting, G.B. Dias, A. Luhur, A.C. Zelhof, C.M. Bergman (2021) Transposable element profiles reveal cell line identity and loss of heterozygosity in Drosophila cell culture. Genetics 219(2):iyab113