biothings_schema
biothings_schema is a Python package for the creation, extension and exploration of the schemas defined using the schema.org standard.
Feature Support
In [1]: from biothings_schema import Schema
In [2]: schema_file_path = "https://raw.githubusercontent.com/data2health/schemas/biothings/biothings/biothings_curie_kevin.jsonld"
In [3]: se = Schema(schema_file_path)
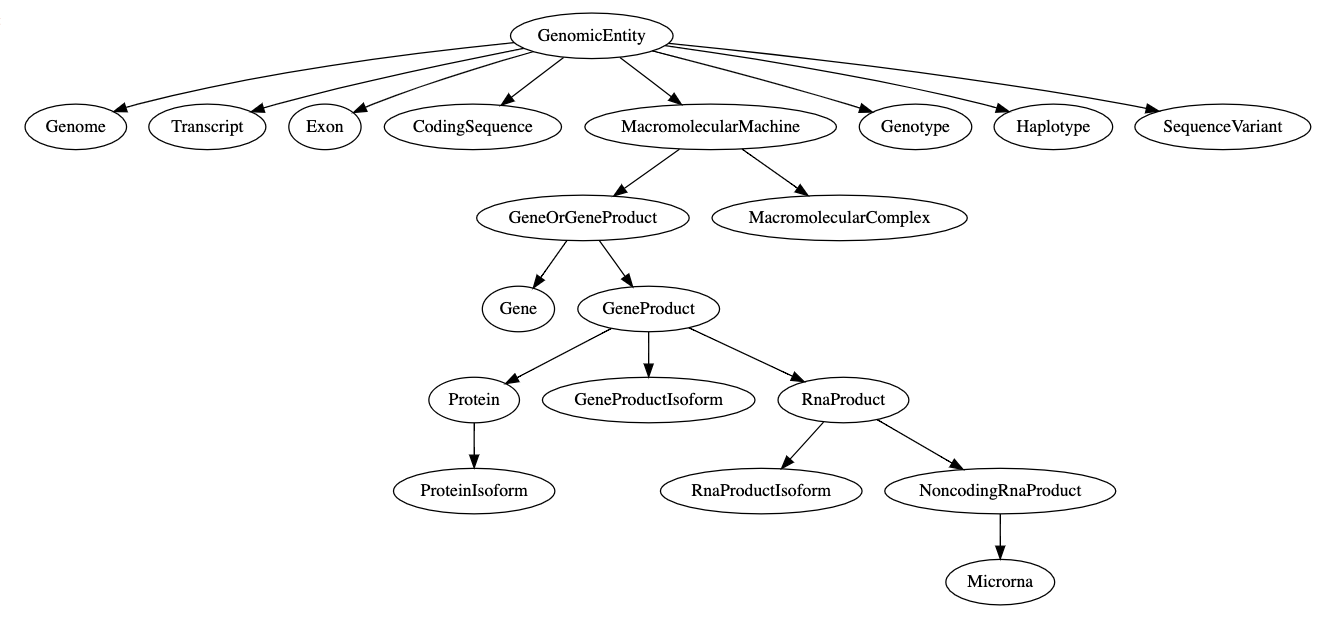
In [4]: scls = se.get_class("Gene")
In [5]: scls.parent_classes
Out [5]:
[[<SchemaClass "Thing">,
<SchemaClass "BiologicalEntity">,
<SchemaClass "MolecularEntity">,
<SchemaClass "GenomicEntity">,
<SchemaClass "MacromolecularMachine">,
<SchemaClass "GeneOrGeneProduct">]]
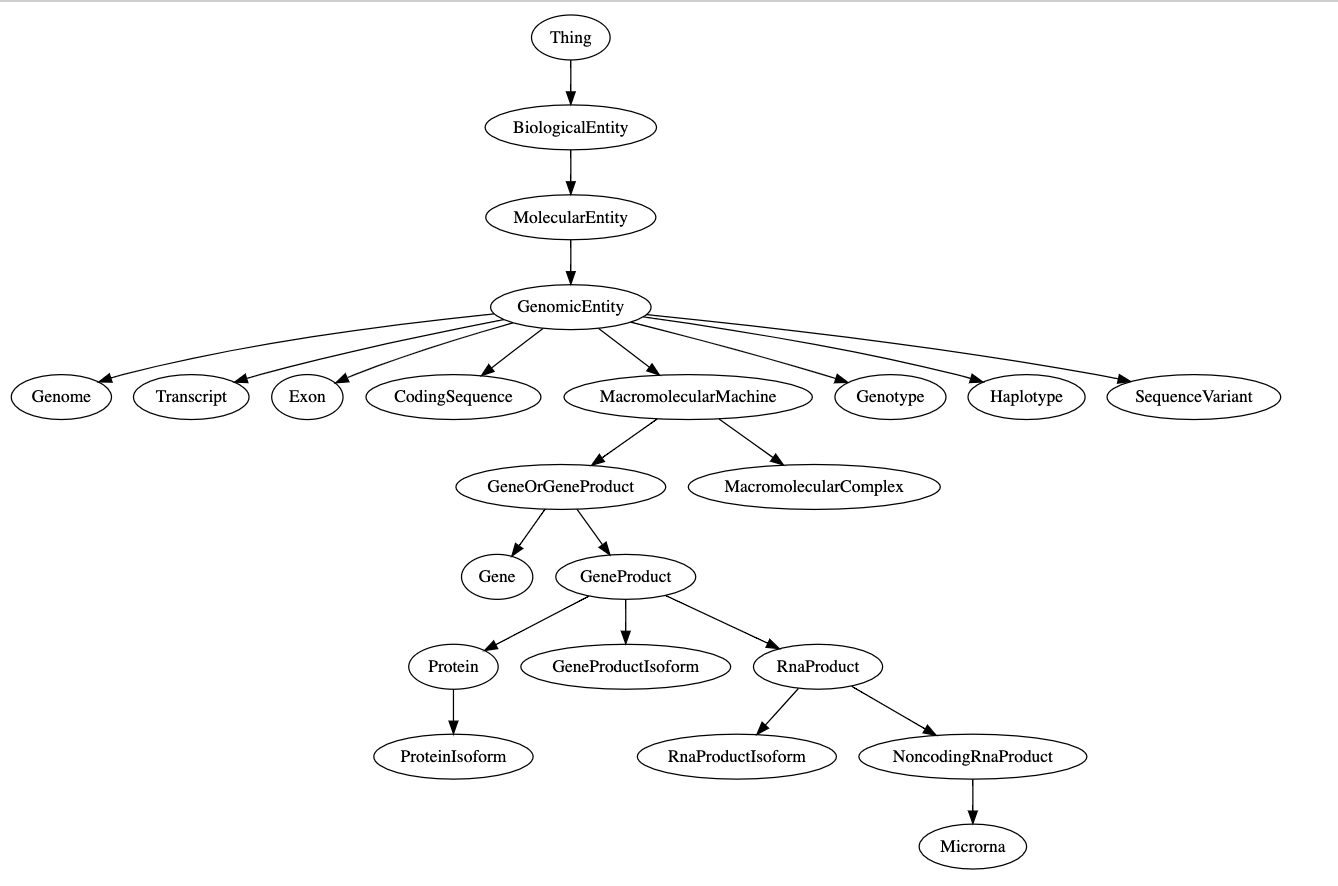
In [6]: scls = se.get_class("MolecularEntity")
In [7]: scls.descendant_classes
Out [7]:
[<SchemaClass "Genome">,
<SchemaClass "RnaProductIsoform">,
<SchemaClass "GeneProduct">,
<SchemaClass "Metabolite">,
<SchemaClass "Protein">,
<SchemaClass "MacromolecularComplex">,
<SchemaClass "Microrna">,
<SchemaClass "Exon">,
<SchemaClass "RnaProduct">,
<SchemaClass "CodingSequence">,
<SchemaClass "GeneProductIsoform">,
<SchemaClass "GeneFamily">,
<SchemaClass "Drug">,
<SchemaClass "Haplotype">,
<SchemaClass "ChemicalSubstance">,
<SchemaClass "Genotype">,
<SchemaClass "ProteinIsoform">,
<SchemaClass "NoncodingRnaProduct">,
<SchemaClass "GeneOrGeneProduct">,
<SchemaClass "Gene">,
<SchemaClass "MacromolecularMachine">,
<SchemaClass "Transcript">,
<SchemaClass "SequenceVariant">,
<SchemaClass "GenomicEntity">]In [8]: scls = se.get_class("Gene")
In [9]: scls.list_properties(group_by_class=False)
Out [9]:
[<SchemaProperty "mgi"">,
<SchemaProperty "pombase"">,
<SchemaProperty "rgd"">,
<SchemaProperty "umls"">,
<SchemaProperty "omim"">,
<SchemaProperty "hasTranscript"">,
<SchemaProperty "dictybase"">,
<SchemaProperty "geneAssociatedWithCondition"">,
<SchemaProperty "flybase"">,
<SchemaProperty "hasGeneProduct"">,
<SchemaProperty "pharos"">,
<SchemaProperty "pharmgkb"">,
<SchemaProperty "unigene"">,
<SchemaProperty "symbol"">,
<SchemaProperty "zfin"">,
<SchemaProperty "entrez"">,
<SchemaProperty "inTaxon"">,
<SchemaProperty "hgnc"">,
<SchemaProperty "geneticallyInteractsWith"">,
<SchemaProperty "tair"">,
<SchemaProperty "sgd"">]-
Edit/Extend your schema
For all features supported by biothings_schema as well as their usage, please see detailed jupyter notebook demo.
Installation
To install biothings_schema, simply use pip:
$ pip install git+https://github.com/biothings/biothings_schema.py#egg=biothings_schemaDocumentation
Fantastic documentation is available at https://biothings-schemapy.readthedocs.io/en/latest/.
The Contributor Guide
If you want to contribute to the project, please refer to the contribution guidelines here