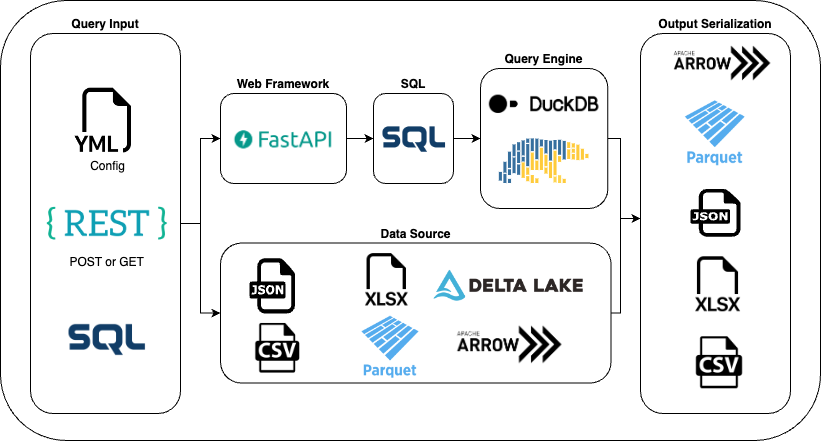
The BMS LakeAPI
A FastAPI plugin that allows you to expose your data lake as an API, allowing several output formats such as Parquet, Csv, Json, Excel, ...
The LakeAPI also includes a minimal security layer for convenience (Basic Auth), but you can also bring your own.
Unlike roapi, we intentionally do not expose most SQL by default, but limit the possible queries with a config. This makes it easy for you to control what happens to your data. If you want the SQL endpoint, you can enable it. And because we built LakeAPI on the shoulders of giants like FastAPI. A lot of things like documentation and authentication are built in and we don't have to reinvent the wheel.
To run the application with the default config, just do it:
app = FastAPI()
bmsdna.lakeapi.init_lakeapi(app)To adjust the config, you can do like this:
import dataclasses
import bmsdna.lakeapi
def_cfg = bmsdna.lakeapi.get_default_config() # Get default startup config
cfg = dataclasses.replace(def_cfg, enable_sql_endpoint=True, data_path="tests/data") # Use dataclasses.replace to set the properties you want
sti = bmsdna.lakeapi.init_lakeapi(app, cfg, "config_test.yml") # Enable it. The first parameter is the FastAPI instance, the 2nd one is the basic config and the third one the config of the tablesInstallation
Pypi Package bmsdna-lakeapi can be installed like any python package : pip install bmsdna-lakeapi
Basic Idea
Based on a YAML configuration and the data source, LakeAPI will automatically generate GET and/or POST endpoints.
Calling the endpoint turns the query into an SQL statement that can be executed with the engine of your choice (duckdb or polars).
The result is then seralised into the requested format (Json, CSV, Arrow etc).
This makes it super easy to distribute your data lake data to other systems. We use it internally to feed data to MS SQL Server, SQLite, Streamlit and Web Apps. You can host it wherever you want, we use Azure Websites which works fine even for very large data amounts.
OpenAPI
Of course everything works with OpenAPI and FastAPI. This means you can add other FastAPI routes, you can use the /docs and /redoc endpoints.
So everything will be fully documented automatically, which is really cool. 🔥🔥
Engine
DuckDB is the default query engine. Polars is also supported, but lack some features. The query engine can be specified at the route level and at the query level with the hidden parameter $engine="duckdb|polars". If you want polars, add the required extra.
At the moment, DuckDB seems to have an advantage and performs the best. Also features like full text search are only available with DuckDB.
A note on Deltalake Support
Deltalake is supported WITH Column Mapping, thanks to deltalake2db in Polars and DuckDB. Deletion Vectors are not supported yet.
Default Security
By Default, Basic Authentication is enabled. To add a user, simply run add_lakeapi_user YOURUSERNAME --yaml-file config.yml. This will add the user to your config yaml (argon2 encrypted).
The generated Password is printed. If you do not want this logic, you can overwrite the username_retriver of the Default Config
Standalone Mode
If you just want to run this thing, you can run it with a webserver:
Uvicorn: uvicorn bmsdna.lakeapi.standalone:app --host 0.0.0.0 --port 8080
Gunicorn: gunicorn bmsdna.lakeapi.standalone:app --workers 4 --worker-class uvicorn.workers.UvicornWorker --bind 0.0.0.0:80
Of course you need to adjust your http options as needed. Also, you need to pip install uvicorn/gunicorn
You can still use environment variables for configuration
Environment Variables
- CONFIG_PATH: The path of the config file, defaults to
config.yml. If you want to split the config, you can specify a folder, too - DATA_PATH: The path of the data files, defaults to
data. Paths inconfig.ymlare relative to DATA_PATH - ENABLE_SQL_ENDPOINT: Set this to 1 to enable the SQL Endpoint
Config File
The application by default relies on a Config file beeing present at the root of your project that's call config.yml.
The config file looks something like this, see also our test yaml:
tables:
- name: fruits
tag: test
version: 1
api_method:
- get
- post
params:
- name: cars
operators:
- "="
- in
- name: fruits
operators:
- "="
- in
datasource:
uri: delta/fruits
file_type: delta
- name: fruits_partition
tag: test
version: 1
api_method:
- get
- post
params:
- name: cars
operators:
- "="
- in
- name: fruits
operators:
- "="
- in
- name: pk
combi:
- fruits
- cars
- name: combi
combi:
- fruits
- cars
datasource:
uri: delta/fruits_partition
file_type: delta
select:
- name: A
- name: fruits
- name: B
- name: cars
- name: fake_delta
tag: test
version: 1
allow_get_all_pages: true
api_method:
- get
- post
params:
- name: name
operators:
- "="
- name: name1
operators:
- "="
datasource:
uri: delta/fake
file_type: delta
# use duckdb database as file
- name: fruits_duck
tag: test
version: 1
api_method:
- get
- post
datasource:
uri: duckdb/fruits.db
file_type: duckdb
table_name: fruits
params:
- name: fruits
operators:
- "="
- in
- name: cars
operators:
- "="
- "in"
- name: fake_delta_partition
tag: test
version: 1
allow_get_all_pages: true
api_method:
- get
- post
params:
- name: name
operators:
- "="
- name: name1
operators:
- "="
datasource:
uri: delta/fake
file_type: delta
- name: "*" # We're lazy and want to expose all in that folder. Name MUST be * and nothing else
tag: startest
version: 1
api_method:
- post
datasource:
uri: startest/* # Uri MUST end with /*
file_type: delta
- name: fruits # But we want to overwrite this one
tag: startest
version: 1
api_method:
- get
datasource:
uri: startest/fruits/${IN_URI_YOU_CAN_HAVE_ENVIRONMENT_VARIABLES}
file_type: delta
- name: mssql_department
tag: mssql
api_method: get
engine: odbc #requires the odbc extra
datasource:
uri: DRIVER={ODBC Driver 17 for SQL Server};SERVER=127.0.0.1,1439;ENCRYPT=yes;TrustServerCertificate=Yes;UID=sa;PWD=${MY_SQL_PWD};Database=AdventureWorks
table_name: "HumanResources.Department"
params:
- GroupNamePartitioning for awesome performance
To use partitions, you can either
- Partition by a column that you filter on. Obviously
- partition on a special column called
columnname_md5_prefix_2, which means that you're partitioning on the first two characters of your hex-coded md5 hash. of the hexadecimal md5 hash. If you now filter bycolumnname, this will greatly reduce the number of files you search for. The number of characters used is up to you, we found two to be useful - partition on a special column called
columnname_md5_mod_NRPARTITIONS, where your partition value isstr(int(hashlib.md5(COLUMNNAME).hexdigest(), 16) % NRPARTITIONS). This might look a bit complicated, but it's not that hard :) You're just doing a modulo on your md5 hash, which allows you to which allows you to set the exact number of partitions. Filtering is still done correctly oncolumnname.
Why partition by MD5 hash? Imagine you have a product id where most id's start with a 1 and some newer ones start with a 2. Most of the data will be in the first partition. If you use an MD5 hash, the data will be spread evenly across the partitions.
With this hack you can get sub-second results on very large data. 🚀🚀
You need to use deltalake to use partitions, and you only need str partition columns for now.
Z-ordering can also help a lot :).
Even more features
- Built-in paging, you can use limit/offset to control what you get
- Full-text search using DuckDB's full-text search feature
- jsonify_complex parameter to convert structs/lists to json, the client cannot handle structs/lists
- Metadata endpoints to retrieve data types, string lengths and more
- Easily expose entire folders by using a "*" wildcard in both the name and the datasource.uri config, see example in the config above
- Config in delta table as json, by using the lakeapi.config Table property. Example in the tests. This allows to have the config for the lakeapi along with your data
- Good test coverage
ODBC Support / Sqlite Support
If you want to use ODBC , install the required extra (named odbc).
ODBC Tables cannot be queried through the sql endpoint for now because of Security Implications. We might allow this later if needed via explicit flag.
ODBC is tested against MS SQL Server using ODBC Driver 17 for MS SQL Server
To use Sqlite, you will need to install adbc_driver_sqlite yourself (pip install adbc_driver_sqlite).
Azure Support
Azure Support is tested and supported. S3/GCP could work in theory but is not tested. Supported configs are taken from object_store create and fsspec:
- https://docs.rs/object_store/latest/object_store/azure/enum.AzureConfigKey.html#variants
- https://github.com/fsspec/adlfs#setting-credentials
Supported settings are only those which are supported by BOTH projects. In practice you can currently use:
- account_name
- account_key
- use_emulator (we add that one for fsspec)
- anon (fsspec/adlfs for anonymous). Adlfs default this to true, we default to false. Set to true if you really want to.
If you want to use Azure Default Credential, just specify account_name without any further option.
Further projects
- lakeapi2sql Allows you to read from lake api and write to MS SQL Server
Work in progress
Please note that this is a work in progress, changes will be made and things may break. Especially at this early stage.
We recommend only using the export from bmsdna.lakeapi for now and not to use the submodules.
Limitations
- You can currently only serve local files. We might add support for
fsspeclater on. You can still mount your data if you use Linux, which is what we do in production
Contribution
Feel free to contribute, report bugs or request enhancements.