
Data processing for image-based profiling
Pycytominer is a suite of common functions used to process high dimensional readouts from high-throughput cell experiments. The tool is most often used for processing data through the following pipeline:
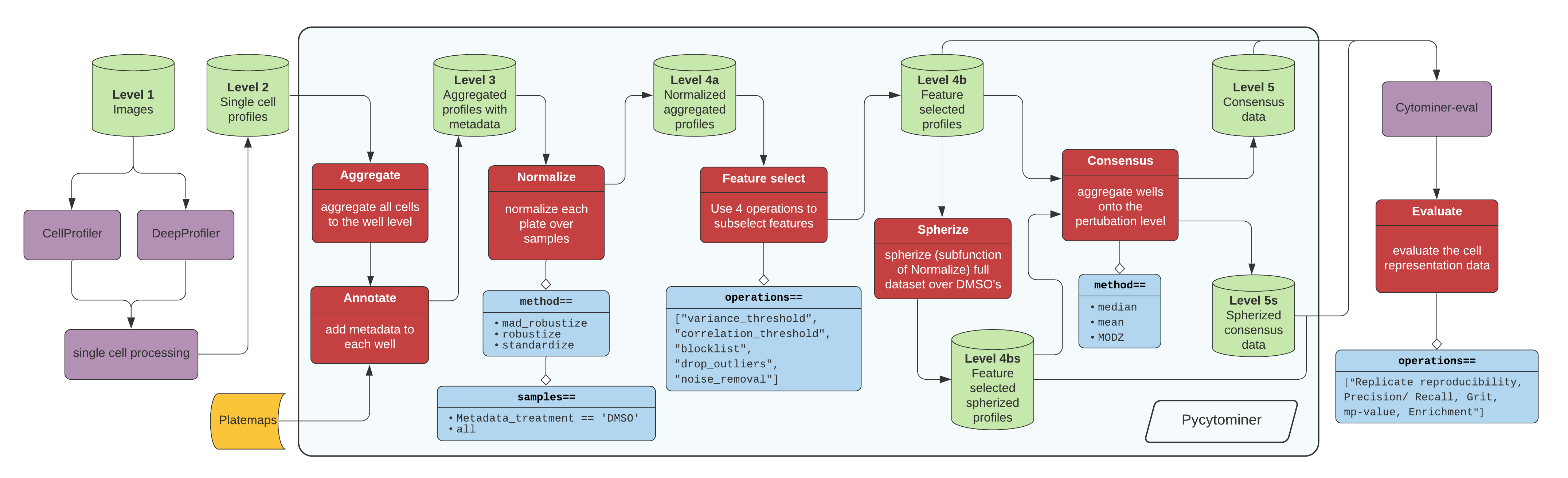
Figure 1. The standard image-based profiling experiment and the role of Pycytominer. (A) In the experimental phase, a scientist plates cells, often perturbing them with chemical or genetic agents and performs microscopy imaging. In image analysis, using CellProfiler for example, a scientist applies several data processing steps to generate image-based profiles. In addition, scientists can apply a more flexible approach by using deep learning models, such as DeepProfiler, to generate image-based profiles. (B) Pycytominer performs image-based profiling to process morphology features and make them ready for downstream analyses. (C) Pycytominer performs five fundamental functions, each implemented with a simple and intuitive API. Each function enables a user to implement various methods for executing operations.
Click here for high resolution pipeline image
Image data flow from a microscope to cell segmentation and feature extraction tools (e.g. CellProfiler or DeepProfiler) (Figure 1A). From here, additional single cell processing tools curate the single cell readouts into a form manageable for pycytominer input. For CellProfiler, we use cytominer-database or CytoTable. For DeepProfiler, we include single cell processing tools in pycytominer.cyto_utils.
Next, Pycytominer performs reproducible image-based profiling (Figure 1B). The Pycytominer API consists of five key steps (Figure 1C). The outputs generated by Pycytominer are utilized for downstream analysis, which includes machine learning models and statistical testing to derive biological insights.
The best way to communicate with us is through GitHub Issues, where we are able to discuss and troubleshoot topics related to pycytominer.
Please see our CONTRIBUTING.md for details about communicating possible bugs, new features, or other information.
Installation
You can install pycytominer using the following platforms.
This project follows a <major>.<minor>.<patch> semantic versioning scheme which is used for every release with small variations per platform.
pip (link):
# install pycyotminer from PyPI
pip install pycytominerconda (link):
# install pycytominer from conda-forge
conda install -c conda-forge pycytominerDocker Hub (link):
Container images of pycytominer are made available through Docker Hub.
These images follow a tagging scheme that extends our release sematic versioning which may be found within our CONTRIBUTING.md Docker Hub Image Releases documentation.
# pull the latest pycytominer image and run a module
docker run --platform=linux/amd64 cytomining/pycytominer:latest python -m pycytominer.<modules go here>
# pull a commit-based version of pycytominer (b1bb292) and run an interactive bash session within the container
docker run -it --platform=linux/amd64 cytomining/pycytominer:pycytominer-1.1.0.post16.dev0_b1bb292 bash
# pull a scheduled update of pycytominer, map the present working directory to /opt within the container, and run a python script.
docker run -v $PWD:/opt --platform=linux/amd64 cytomining/pycytominer:pycytominer-1.1.0.post16.dev0_b1bb292_240417 python /opt/script.pyFrameworks
Pycytominer is primarily built on top of pandas, also using aspects of SQLAlchemy, sklearn, and pyarrow.
Pycytominer currently supports parquet and compressed text file (e.g. .csv.gz) i/o.
CellProfiler support
Currently, Pycytominer fully supports data generated by CellProfiler, adhering defaults to its specific data structure and naming conventions.
CellProfiler-generated image-based profiles typically consist of two main components:
- Metadata features: This section contains information about the experiment, such as plate ID, well position, incubation time, perturbation type, and other relevant experimental details. These feature names are prefixed with
Metadata_, indicating that the data in these columns contain metadata information. - Morphology features: These are the quantified morphological features prefixed with the default compartments (
Cells_,Cytoplasm_, andNuclei_). Pycytominer also supports non-default compartment names (e.g.,Mito_).
Note, pycytominer.cyto_utils.cells.SingleCells() contains code designed to interact with single-cell SQLite files exported from CellProfiler.
Processing capabilities for SQLite files depends on SQLite file size and your available computational resources (for ex. memory and CPU).
Handling inputs from other image analysis tools (other than CellProfiler)
Pycytominer also supports processing of raw morphological features from image analysis tools beyond CellProfiler. These tools include In Carta, Harmony, and others. Using Pycytominer with these tools requires minor modifications to function arguments, and we encourage these users to pay particularly close attention to individual function documentation.
For example, to resolve potential feature issues in the normalize() function, you must manually specify the morphological features using the features parameter.
The features parameter is also available in other key steps, such as aggregate and feature_select.
If you are using Pycytominer with these other tools, please file an issue to reach out. We'd love to hear from you so that we can learn how to best support broad and multiple use-cases.
API
Pycytominer has five major processing functions:
- Aggregate - Average single-cell profiles based on metadata information (most often "well").
- Annotate - Append metadata (most often from the platemap file) to the feature profile
- Normalize - Transform input feature data into consistent distributions
- Feature select - Exclude non-informative or redundant features
- Consensus - Average aggregated profiles by replicates to form a "consensus signature"
The API is consistent for each of these functions:
# Each function takes as input a pandas DataFrame or file path
# and transforms the input data based on the provided options and methods
df = function(
profiles_or_path,
features,
samples,
method,
output_file,
additional_options...
)Each processing function has unique arguments, see our documentation for more details.
Usage
The default way to use pycytominer is within python scripts, and using pycytominer is simple and fun.
The example below demonstrates how to perform normalization with a dataset generated by CellProfiler.
# Real world example
import pandas as pd
import pycytominer
commit = "da8ae6a3bc103346095d61b4ee02f08fc85a5d98"
url = f"https://media.githubusercontent.com/media/broadinstitute/lincs-cell-painting/{commit}/profiles/2016_04_01_a549_48hr_batch1/SQ00014812/SQ00014812_augmented.csv.gz"
df = pd.read_csv(url)
normalized_df = pycytominer.normalize(
profiles=df,
method="standardize",
samples="Metadata_broad_sample == 'DMSO'"
)Pipeline orchestration
Pycytominer is a collection of different functions with no explicit link between steps. However, some options exist to use pycytominer within a pipeline framework.
| Project | Format | Environment | pycytominer usage |
|---|---|---|---|
| Profiling-recipe | yaml | agnostic | full pipeline support |
| CellProfiler-on-Terra | WDL | google cloud / Terra | single-cell aggregation |
| CytoSnake | snakemake | agnostic | full pipeline support |
A separate project called AuSPICES offers pipeline support up to image feature extraction.
Other functionality
Pycytominer was written with a goal of processing any high-throughput image-based profiling data. However, the initial use case was developed for processing image-based profiling experiments specifically. And, more specifically than that, image-based profiling readouts from CellProfiler measurements from Cell Painting data.
Therefore, we have included some custom tools in pycytominer/cyto_utils that provides other functionality:
CellProfiler CSV collation
If running your images on a cluster, unless you have a MySQL or similar large database set up then you will likely end up with lots of different folders from the different cluster runs (often one per well or one per site), each one containing an Image.csv, Nuclei.csv, etc.
In order to look at full plates, therefore, we first need to collate all of these CSVs into a single file (currently SQLite) per plate.
We currently do this with a library called cytominer-database.
If you want to perform this data collation inside pycytominer using the cyto_utils function collate (and/or you want to be able to run the tests and have them all pass!), you will need cytominer-database==0.3.4; this will change your installation commands slightly:
# Example for general case commit:
pip install "pycytominer[collate]"
# Example for specific commit:
pip install "pycytominer[collate] @ git+https://github.com/cytomining/pycytominer@77d93a3a551a438799a97ba57d49b19de0a293ab"If using pycytominer in a conda environment, in order to run collate.py, you will also want to make sure to add cytominer-database=0.3.4 to your list of dependencies.
Creating a cell locations lookup table
The CellLocation class offers a convenient way to augment a LoadData file with X,Y locations of cells in each image.
The locations information is obtained from a single cell SQLite file.
To use this functionality, you will need to modify your installation command, similar to above:
# Example for general case commit:
pip install "pycytominer[cell_locations]"Example using this functionality:
metadata_input="s3://cellpainting-gallery/test-cpg0016-jump/source_4/workspace/load_data_csv/2021_08_23_Batch12/BR00126114/test_BR00126114_load_data_with_illum.parquet"
single_single_cell_input="s3://cellpainting-gallery/test-cpg0016-jump/source_4/workspace/backend/2021_08_23_Batch12/BR00126114/test_BR00126114.sqlite"
augmented_metadata_output="~/Desktop/load_data_with_illum_and_cell_location_subset.parquet"
python \
-m pycytominer.cyto_utils.cell_locations_cmd \
--metadata_input ${metadata_input} \
--single_cell_input ${single_single_cell_input} \
--augmented_metadata_output ${augmented_metadata_output} \
add_cell_location
# Check the output
python -c "import pandas as pd; print(pd.read_parquet('${augmented_metadata_output}').head())"
# It should look something like this (depends on the width of your terminal):
# Metadata_Plate Metadata_Well Metadata_Site ... PathName_OrigRNA ImageNumber CellCenters
# 0 BR00126114 A01 1 ... s3://cellpainting-gallery/cpg0016-jump/source_... 1 [{'Nuclei_Location_Center_X': 943.512129380054...
# 1 BR00126114 A01 2 ... s3://cellpainting-gallery/cpg0016-jump/source_... 2 [{'Nuclei_Location_Center_X': 29.9516027655562...Generating a GCT file for morpheus
The software morpheus enables profile visualization in the form of interactive heatmaps.
Pycytominer can convert profiles into a .gct file for drag-and-drop input into morpheus.
# Real world example
import pandas as pd
import pycytominer
commit = "da8ae6a3bc103346095d61b4ee02f08fc85a5d98"
plate = "SQ00014812"
url = f"https://media.githubusercontent.com/media/broadinstitute/lincs-cell-painting/{commit}/profiles/2016_04_01_a549_48hr_batch1/{plate}/{plate}_normalized_feature_select.csv.gz"
df = pd.read_csv(url)
output_file = f"{plate}.gct"
pycytominer.cyto_utils.write_gct(
profiles=df,
output_file=output_file
)Citing Pycytominer
If you have used pycytominer in your project, please use the citation below.
You can also find the citation in the 'cite this repository' link at the top right under about section.
APA:
Serrano, E., Chandrasekaran, N., Bunten, D., Brewer, K., Tomkinson, J., Kern, R., Bornholdt, M., Fleming, S., Pei, R., Arevalo, J., Tsang, H., Rubinetti, V., Tromans-Coia, C., Becker, T., Weisbart, E., Bunne, C., Kalinin, A. A., Senft, R., Taylor, S. J., Jamali, N., Adeboye, A., Abbasi, H. S., Goodman, A., Caicedo, J., Carpenter, A. E., Cimini, B. A., Singh, S., & Way, G. P. Reproducible image-based profiling with Pycytominer. https://doi.org/10.48550/arXiv.2311.13417
