CDMConnector
Are you using the tidyverse with an OMOP Common Data Model?
Interact with your CDM in a pipe-friendly way with CDMConnector.
- Quickly connect to your CDM and start exploring.
- Build data analysis pipelines using familiar dplyr verbs.
- Easily extract subsets of CDM data from a database.
Overview
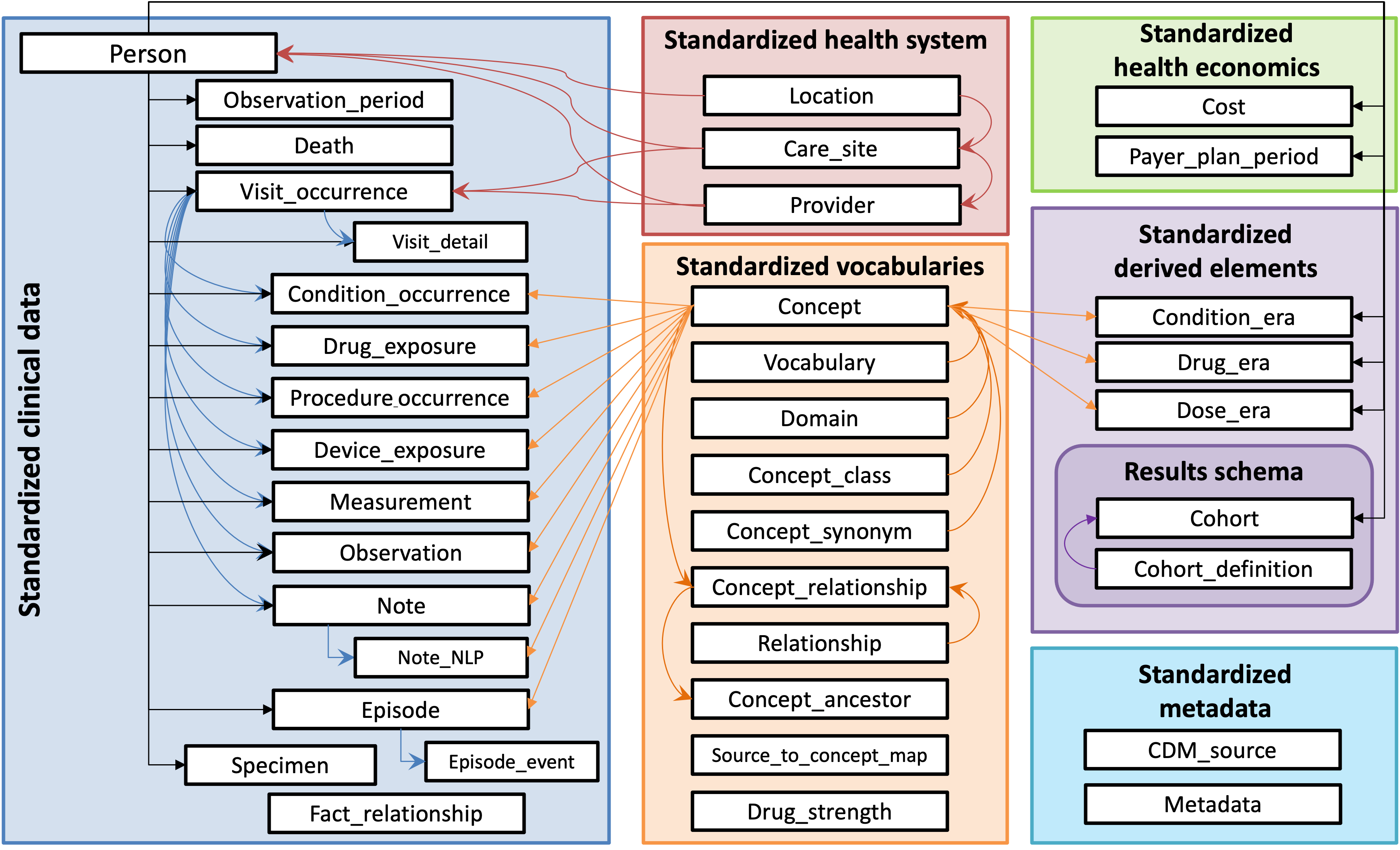
CDMConnector introduces a single R object that represents an OMOP CDM relational database inspired by the dm, DatabaseConnector, and Andromeda packages. The cdm objects encapsulate references to OMOP CDM tables in a remote RDBMS as well as metadata necessary for interacting with a CDM, allowing for dplyr style data analysis pipelines and interactive data exploration.
Features
CDMConnector is meant to be the entry point for composable tidyverse
style data analysis operations on an OMOP CDM. A cdm_reference object
behaves like a named list of tables.
- Quickly create a list of references to a subset of CDM tables
- Store connection information for later use inside functions
- Use any DBI driver back-end with the OMOP CDM
See Getting started for more details.
Installation
CDMConnector can be installed from CRAN:
install.packages("CDMConnector")The development version can be installed from GitHub:
# install.packages("devtools")
devtools::install_github("darwin-eu/CDMConnector")Usage
Create a cdm reference from any DBI connection to a database containing OMOP CDM tables. Use the cdm_schema argument to point to a particular schema in your database that contains your OMOP CDM tables and the write_schema to specify the schema where results tables can be created, and use cdm_name to provide a name for the database.
library(CDMConnector)
con <- DBI::dbConnect(duckdb::duckdb(dbdir = eunomia_dir()))
cdm <- cdm_from_con(con = con,
cdm_schema = "main",
write_schema = "main",
cdm_name = "my_duckdb_database")## Note: method with signature 'DBIConnection#Id' chosen for function 'dbExistsTable',
## target signature 'duckdb_connection#Id'.
## "duckdb_connection#ANY" would also be validA cdm_reference is a named list of table references:
library(dplyr)
names(cdm)## [1] "person" "observation_period" "visit_occurrence"
## [4] "visit_detail" "condition_occurrence" "drug_exposure"
## [7] "procedure_occurrence" "device_exposure" "measurement"
## [10] "observation" "death" "note"
## [13] "note_nlp" "specimen" "fact_relationship"
## [16] "location" "care_site" "provider"
## [19] "payer_plan_period" "cost" "drug_era"
## [22] "dose_era" "condition_era" "metadata"
## [25] "cdm_source" "concept" "vocabulary"
## [28] "domain" "concept_class" "concept_relationship"
## [31] "relationship" "concept_synonym" "concept_ancestor"
## [34] "source_to_concept_map" "drug_strength"Use dplyr verbs with the table references.
cdm$person %>%
tally()## # Source: SQL [1 x 1]
## # Database: DuckDB v1.1.2 [root@Darwin 23.1.0:R 4.3.3//private/var/folders/2j/8z0yfn1j69q8sxjc7vj9yhz40000gp/T/RtmpDw9JTb/fileeea2255bd10b.duckdb]
## n
## <dbl>
## 1 2694Compose operations with the pipe.
cdm$condition_era %>%
left_join(cdm$concept, by = c("condition_concept_id" = "concept_id")) %>%
count(top_conditions = concept_name, sort = TRUE)## # Source: SQL [?? x 2]
## # Database: DuckDB v1.1.2 [root@Darwin 23.1.0:R 4.3.3//private/var/folders/2j/8z0yfn1j69q8sxjc7vj9yhz40000gp/T/RtmpDw9JTb/fileeea2255bd10b.duckdb]
## # Ordered by: desc(n)
## top_conditions n
## <chr> <dbl>
## 1 Viral sinusitis 17268
## 2 Acute viral pharyngitis 10217
## 3 Acute bronchitis 8184
## 4 Otitis media 3561
## 5 Osteoarthritis 2694
## 6 Streptococcal sore throat 2656
## 7 Sprain of ankle 1915
## 8 Concussion with no loss of consciousness 1013
## 9 Sinusitis 1001
## 10 Acute bacterial sinusitis 939
## # ℹ more rowsAnd much more besides. See vignettes for further explanations on how to create database connections, make a cdm reference, and start analysing your data.
Getting help
If you encounter a clear bug, please file an issue with a minimal reproducible example on GitHub.
Citation
## To cite package 'CDMConnector' in publications use:
##
## Black A, Gorbachev A, Burn E, Catala Sabate M (????). _CDMConnector:
## Connect to an OMOP Common Data Model_. R package version 1.6.0,
## https://github.com/darwin-eu/CDMConnector,
## <https://darwin-eu.github.io/CDMConnector/>.
##
## A BibTeX entry for LaTeX users is
##
## @Manual{,
## title = {CDMConnector: Connect to an OMOP Common Data Model},
## author = {Adam Black and Artem Gorbachev and Edward Burn and Marti {Catala Sabate}},
## note = {R package version 1.6.0, https://github.com/darwin-eu/CDMConnector},
## url = {https://darwin-eu.github.io/CDMConnector/},
## }License: Apache 2.0