CRISPRCasFinder
CRISPRCasFinder is an updated, improved, and integrated version of CRISPRFinder and CasFinder.

References/Citations
If you use this software, please cite:
-
Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007 Jul;35(Web Server issue):W52-7. DOI: https://doi.org/10.1093/nar/gkm360 PMID:17537822
-
Abby SS, Néron B, Ménager H, Touchon M, Rocha EP. MacSyFinder: a program to mine genomes for molecular systems with an application to CRISPR-Cas systems. PLoS One. 2014 Oct 17;9(10):e110726. DOI: https://doi.org/10.1371/journal.pone.0110726 PMID:25330359
-
Couvin D, Bernheim A, Toffano-Nioche C, Touchon M, Michalik J, Néron B, Rocha EPC, Vergnaud G, Gautheret D, Pourcel C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018 Jul 2;46(W1):W246-W251. DOI: https://doi.org/10.1093/nar/gky425 PMID:29790974
-
Néron B, Denise R, Coluzzi C, Touchon M, Rocha EPC, Abby SS (2022). MacSyFinder v2: Improved modelling and search engine to identify molecular systems in genomes. bioRxiv DOI: 10.1101/2022.09.02.506364 Link
Further information are available at: https://crisprcas.i2bc.paris-saclay.fr.
Quick Installation
Conda/Bioconda/Mamba
Note that you will first need to install conda/bioconda to run the following commands:
conda env create -f ccf.environment.yml -n crisprcasfinder
conda activate crisprcasfinder
mamba init
mamba activate
mamba install -c bioconda macsyfinder=2.1.2
macsydata install -u CASFinder==3.1.0MacOS
./installer_MAC.shUbuntu
bash installer_UBUNTU.sh
source ~/.profileCentOS
Please first install conda if it is not already installed:
sudo yum -y update
sudo yum -y upgrade
wget https://repo.anaconda.com/miniconda/Miniconda3-latest-Linux-x86_64.sh
bash Miniconda3-latest-Linux-x86_64.sh
export PATH=/path/to/miniconda3/bin/:$PATH
source ~/.bashrc
conda init
conda config --add channels defaults
conda config --add channels bioconda
conda config --add channels conda-forgeNow you can install CRISPRCasFinder as follows:
bash installer_CENTOS.sh
exit #this command could be needed if your command prompt changes
source ~/.bashrcFedora
sudo yum -y update
sudo yum -y upgrade
bash installer_FEDORA.sh
source ~/.bashrcYou can run the command 'perl CRISPRCasFinder.pl -v' to see if everything is OK. You may need to reinstall some Perl's modules (with command: sudo cpanm ...), for example: "sudo cpanm Date::Calc". The notification "Possible precedence issue with control flow operator ..." will not affect results of analysis. For further information, please see the documentation.
To run CRISPRCasFinder in the current directory with example sequence you can type:
perl CRISPRCasFinder.pl -in install_test/sequence.fasta -cas -keepFor further details, please see the documentation.
Documentation
A more complete User Manual is available at the following file : CRISPRCasFinder_Viewer_manual.pdf
Licence
Container
If you want to try CRISPRCasFinder without installing dependencies, The standalone version is also available as a singularity container (hosted on the Download page of the CRISPR-Cas++ portal):
To run the container
Former version of CRISPRCasFinder (v4.2.20)
After downloading the CrisprCasFinder.simg image from the CRISPR-Cas++ Download page, you can for example run the following command (sequence.fasta file must be replaced by your file):
singularity exec -B $PWD CrisprCasFinder.simg perl /usr/local/CRISPRCasFinder/CRISPRCasFinder.pl -so /usr/local/CRISPRCasFinder/sel392v2.so -cf /usr/local/CRISPRCasFinder/CasFinder-2.0.3 -drpt /usr/local/CRISPRCasFinder/supplementary_files/repeatDirection.tsv -rpts /usr/local/CRISPRCasFinder/supplementary_files/Repeat_List.csv -cas -def G -out RES21092020_2 -in sequence.fastaPlease visit the following link for more information about singularity containers: https://www.sylabs.io/docs/
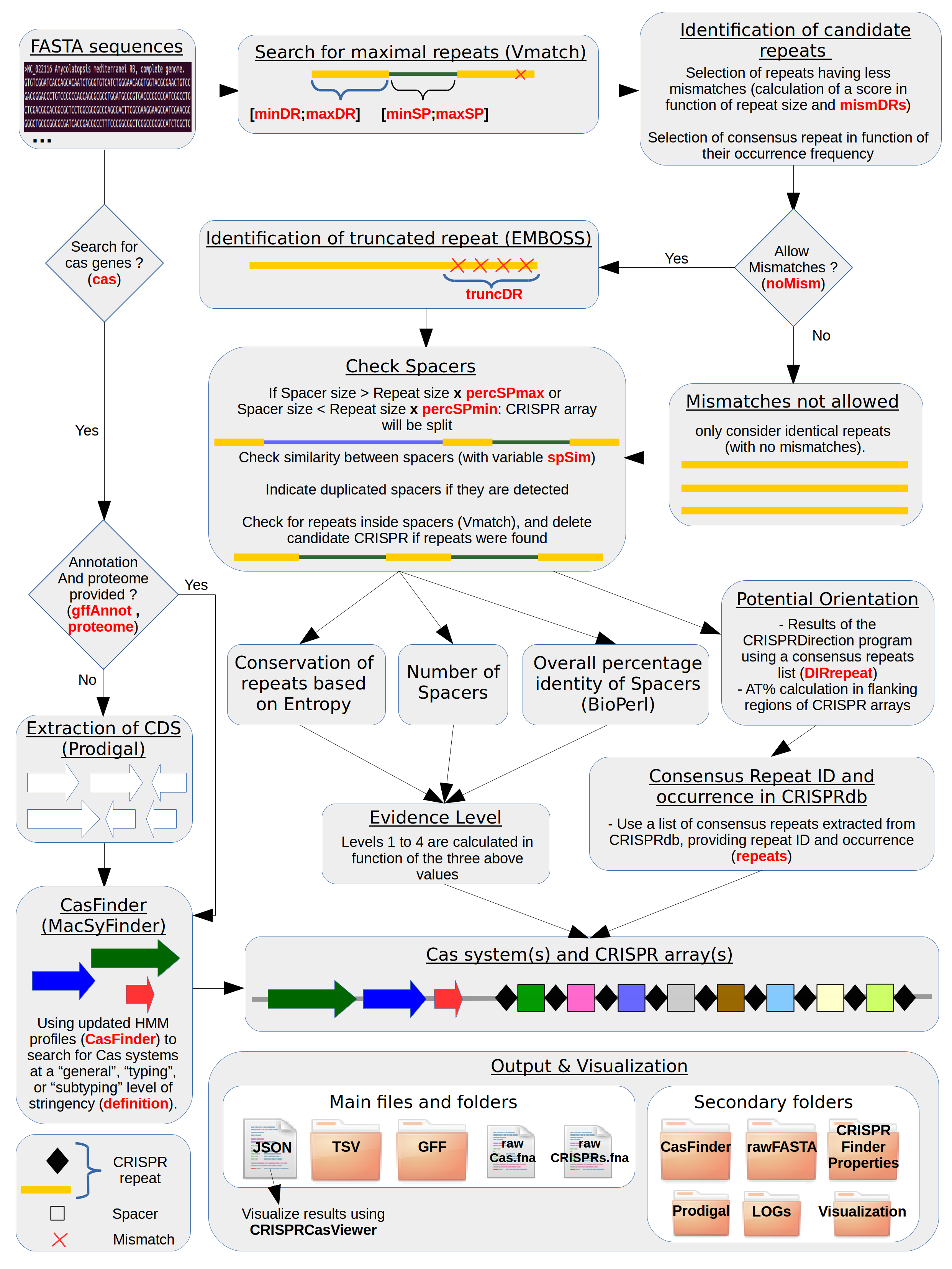
Outline of the CRISPRCasFinder workflow