The eNanoMapper Ontology
The eNanoMapper ontologies aim to provide a comprehensive suite of ontologies for the nanomaterial safety assessment domain (see http://www.enanomapper.net for project information). The full suite of ontologies can be found in a modular file assembled by imports using owl:imports, enanomapper.owl.
The ontology is being developed and maintained by the following EU H2020 projects (see also the below funding info):
- SbD4Nano
- NanoSolveIT (project ended)
- NanoCommons (project ended)
- OpenRiskNet (project ended)
- eNanoMapper (project ended)
Other NanoSafety Cluster projects that have contributed by providing feedback and collaborations include NANoREG, NanoReg2, and GRACIOUS.
External
We import slices from third party ontologies. The Slimmer tool is used to extract the bits we include in the eNanoMapper ontology. The slimmed files can be found in the external folder. The full list of ontologies it includes is:
- Adverse Outcome Pathways Ontology (AOP)
- BioAssay Ontology (BAO)
- Basic Formal Ontology (BFO)
- Cell Culture Ontology (CCONT)
- Chemical Entities of Biological Interest (CHEBI)
- Chemical Information Ontology (CHEMINF)
- Chemical Methods Ontology (CHMO)
- Citation Typing Ontology (CITO)
- Experimental Factor Ontology (EFO)
- Environment Ontology (ENVO)
- FRBR-aligned Bibliographic Ontology (FABIO)
- Gene Ontology (GO)
- Information Artifact Ontology (IAO)
- National Cancer Institute Thesaurus (NCIT)
- NanoParticle Ontology (NPO)
- Ontology of Adverse Events (OAE)
- Ontology of Biological and Clinical Statistics (OBCS)
- Ontology for Biomedical Investigations (OBI)
- Phenotype And Trait Ontology (PATO)
- Semanticscience Integrated Ontology (SIO)
- Uber Anatomy Ontology (UBERON)
- Unit Ontology (UO)
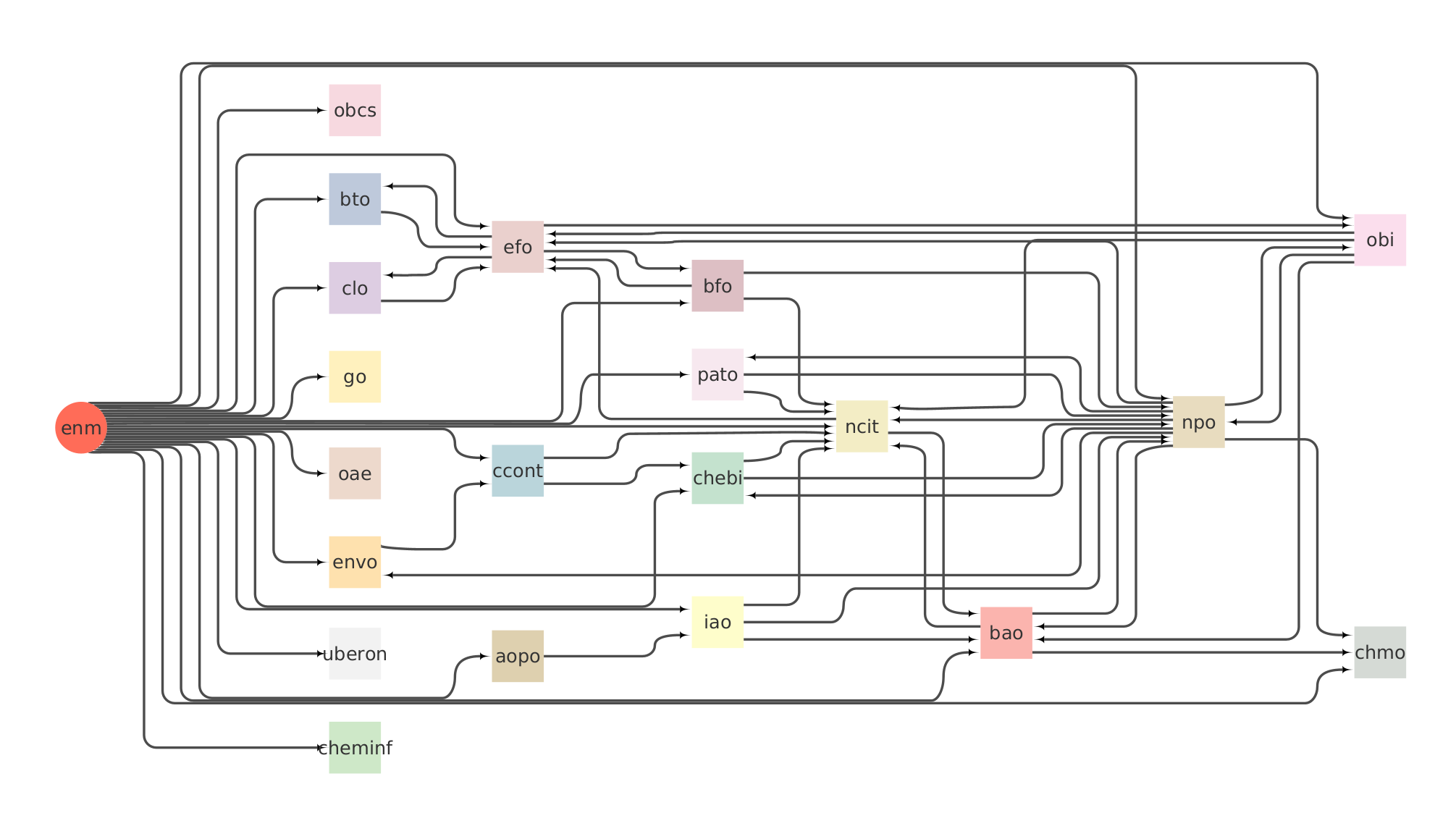

The build of the slims is carried out in this repository through the actions contained in the .github/workflows folder, and the resulting slims of external ontologies are commited and pushed automatically to this repository when the build workflow is run under external-dev.
The configuration file config.yaml is used by the build and test workflows to retrieve ROBOT and Slimmer. There are 3 test workflows:
- On push: test for file integrity (
external,external-dev,internal,internal-dev,config) and performs SPARQL queries onenanomapper-dev.owlto make sure internal terms have not been added at the top of the class hierarchy or directly underentity. - Development tests:
robot diffandrobot reportare performed after each build for QC on the resulting development version of the ontology, with their results being stored as artifacts after each run. - Pre-release tests: ensure the modules are updated and the ontology is satisfiable and has the right top-level class hierarchy before making a release. This is the only test that needs to be dispatched manually.
DOI of Releases
Opening the ontology in Protégé
The stable version can be opened in Protégé with the following step:
- File → Open from URL...
- enter the URL http://enanomapper.github.io/ontologies/enanomapper.owl
The development version is opened in the same way, but with a different URL:
- File → Open from URL...
- enter the URL http://enanomapper.github.io/ontologies/enanomapper-dev.owl
Building and validating the ontology
During (and after) the eNanoMapper project the ontology was autobuilt using scripts on the BiGCaT Jenkins server. After the cyberattack on Maastricht University in 2019, this repository uses GitHub Actions to replicate the Jenkins build process, and adds a full ontology file and customizable workflows for ontology build and QC.
The main OWL file (enanomapper.owl) refers to slimmed versions of external ontologies, complemented with internal
files adding additional terms. The extensions are OWL files themselves and you can load them in Protégé
and use the internal/Makefile to run xmllint on the extensions to see if the OWL
files are well-formed.
The slimming of the external ontologies is done with the Slimmer tool, with these commands (for the BioAssay Ontology):
rm -f *.owl
rm -f *.owl.*
wget -O bao_complete.owl http://www.bioassayontology.org/bao/bao_complete.owl
rm -f bao.props*
rm -f bao.iris*
wget https://raw.githubusercontent.com/enanomapper/ontologies/master/config/bao.props
wget https://raw.githubusercontent.com/enanomapper/ontologies/master/config/bao.iris
java -cp ../Slimmer/target/slimmer-0.0.1-SNAPSHOT-jar-with-dependencies.jar com.github.enanomapper.Slimmer .The bao.props and bao.iris files contain all the information needed to describe which parts of the BAO ontology
is retained in the slimmed version. This is automatically applied for all modules in the build workflow once a month.
Tutorials
Please also check out these tutorials, developed by eNanoMapper, NanoCommons, and OpenRiskNet:
Making Releases
- Check if the metadata in the enanomapper.owl is up to data (e.g. names of people who submitted PRs)
- Copy the internal ontologies in
ontologies/internal-dev/toontologies/internalandontologies/external-dev/toontologies/external - Update for
enanomapper.owlandenanomapper-dev.owl:owl:versionInfoowl:versionIRI(onlyenanomapper.owl)
- Create a folder
releases/{version}and store there a copy of the latestenanomapper.owlfile - Make a git commit with the changes and tag that commit matching the release
- Manually dispatch the
prerelease-testworkflow and make the pertinent fixes if it fails. Repeat the tagged git commit after fixing the issues. - Update the CITATION.cff
titleversiondate-released
- Write markdown for the release with the changes since the previous release
- Release the whole repository in GitHub https://github.com/enanomapper/ontologies/releases
- Update the DOI number for new release: https://zenodo.org/record/260098
Funding
The project has had contributions from various European Commission projects. The eNanoMapper project was funded by the European Union’s Seventh Framework Programme for research, technological development and demonstration (FP7-NMP-2013-SMALL-7) under grant agreement no. 604134. NanoCommons has received funding from European Union Horizon 2020 Programme (H2020) under grant agreement nº 731032. OpenRiskNet is funded by the European Commission within Horizon 2020 EINFRA-22-2016 Programme under grant agreement no. 731075. NanoSolveIt has received funding from European Union Horizon 2020 Programme (H2020) under grant agreement no. 814572. SbD4Nano has received funding from European Union Horizon 2020 Programme (H2020) under grant agreement no. 862195.