SurvLatent ODE
Project Abstract
Effective learning from electronic health records (EHR) data for prediction of clinical outcomes is often challenging due to features recorded at irregular timesteps and loss to follow-up, as well as competing events such as death or disease progression. We propose SurvLatent ODE, a generative time-to-event model using an Ordinary Differential Equation-based Recurrent Neural Networks (ODE-RNN) as an encoder. This model effectively parameterizes dynamics of latent states under irregularly sampled input data and flexibly estimates survival times for multiple competing events. We demonstrate its competitive performance on MIMIC-III and data from the Dana-Farber Cancer Institute (DFCI) for predicting onset of Venous Thromboembolism (VTE) with death as a competing event. SurvLatent ODE outperforms current clinical standards and provides clinically meaningful interpretations.
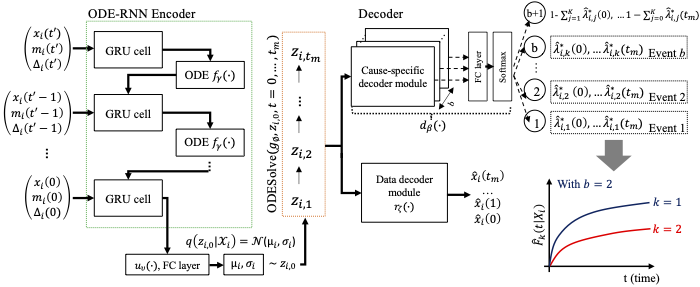
Model Architecture
The SurvLatent ODE model utilizes an ODE-RNN encoder to parameterize a patient-specific temporal trajectory of covariates ($x{i}$(0), $..., x{i}$($t$')) into a latent embedding. This process involves approximating the posterior over the initial latent variable $z_{i,0}$. The initial latent variable is then transformed into a latent trajectory $z_i^{tm} = (z{i,1}, z{i,2}, ..., z{i,tm})$ using a differential equation solver ODESolve($g{\phi}, z_{i,0}, t = 0, ...,tm$). The model's cause-specific decoder modules, a subsequent fully connected layer, and a softmax layer ($m\beta(\cdot)$) then map this latent trajectory to estimate the cause-specific hazard function for each event. For more details, please refer to our paper.
Setting Up the Conda Environment
To run the notebooks and code in this repository, we recommend setting up a Conda environment. This ensures that you have all the necessary dependencies. Follow these steps to create and activate the environment:
-
Install Conda: If you don't have Conda installed, download Miniconda or Anaconda.
-
Create Conda Environment: Create a Conda environment using the
survlatent_ode_conda.ymlfile provided in this repository. Run the following command in your terminal:conda env create -f survlatent_ode_conda.ymlThis command will set up an environment with all the necessary packages as specified in the
survlatent_ode_conda.ymlfile. -
Activate the Environment: Once the environment is created, you can activate it with:
conda activate survlatent_ode
Now you can run the notebooks and scripts within this environment.
Notebook Examples
Our provided notebook examples demonstrate the application of the SurvLatent ODE model using the publicly available Framingham dataset, which aims to understand the etiology of cardiovascular disease.
-
Time to Death Analysis: Notebook example I explores the time to DEATH using the Framingham dataset. This notebook provides insights into the survival analysis and the effectiveness of the model in predicting mortality.
-
Competing Events Analysis: Notebook example II focuses on the time to ANYCHD (Any Coronary Heart Disease), modeling DEATH as a competing event. This example highlights the model's capability in handling complex scenarios where multiple events may influence the outcome.
These notebooks are designed to offer a comprehensive understanding of the model's application in real-world scenarios and its effectiveness in survival analysis.
Citation
@inproceedings{moon2022survlatent,
title={SurvLatent ODE: A Neural ODE based time-to-event model with competing risks for longitudinal data improves cancer-associated Venous Thromboembolism (VTE) prediction},
author={Moon, Intae and Groha, Stefan and Gusev, Alexander},
booktitle={Machine Learning for Healthcare Conference},
pages={800--827},
year={2022},
organization={PMLR}
}