Single-Cell ATAC-seq analysis via Latent feature Extraction

News
2022.06.30 Introduce the highly_variable_genes from scanpy to filter peaks and support for input from multiomics data h5mu
2021.04 A new online integration tool SCALEX on scRNA-seq and scATAC-seq is available!
2021.01.14 Update to compatible with h5ad file and scanpy
Installation
SCALE neural network is implemented in Pytorch framework.
Running SCALE on CUDA is recommended if available.
install from PyPI
pip install scaleinstall latest develop version from GitHub
pip install git+https://github.com/jsxlei/SCALE.gitor download and install
git clone git://github.com/jsxlei/SCALE.git
cd SCALE
python setup.py installInstallation only requires a few minutes.
Quick Start
Input
- h5ad file
- count matrix file:
- row is peak and column is barcode, in txt / tsv (sep="\t") or csv (sep=",") format
- mtx folder contains three files:
- count file: count in mtx format, filename contains key word "count" / "matrix"
- peak file: 1-column of peaks chr_start_end, filename contains key word "peak"
- barcode file: 1-column of barcodes, filename contains key word "barcode"
- h5mu file, e.g. filename.h5mu/atac
Run
SCALE.py -d [input]Output
Output will be saved in the output folder including:
- model.pt: saved model to reproduce results cooperated with option --pretrain
- adata.h5ad: saved data including Leiden cluster assignment, latent feature matrix and UMAP results.
- umap.pdf: visualization of 2d UMAP embeddings of each cell
Imputation
Get binary imputed data in adata.h5ad file using scanpy adata.obsm['binary'] with option --binary (recommended for saving storage)
SCALE.py -d [input] --binary or get numerical imputed data in adata.h5ad file using scanpy adata.obsm['imputed'] with option --impute
SCALE.py -d [input] --imputeUseful options
- [--outdir] or [-o]: save results in a specific folder
- [--embed]: tSNE/UMAP, embed feature by tSNE or UMAP
- [--min_peaks]: filter low quality cells by valid peaks number, default 100
- [--min_cells]: filter low quality peaks by valid cells number, default 3 (previous default is 0.01), now replaced by [--n_feature]
- [--n_feature]: filter peaks by selecting highly variable features, default 100,000; use [--n_feature] -1 to disable.
- [--lr]: modify the initial learning rate, default is 0.002:
- [--max_iter] or [-i]: max iteration number, default is 30000
- [--seed]: random seed for parameter initialization, default is 18
- [--binary]: binarize the imputation values
- [-k]: if cluster number is known
Help
Look for more usage of SCALE
SCALE.py --help Use functions in SCALE packages.
import scale
from scale import *
from scale.plot import *
from scale.utils import *Running time
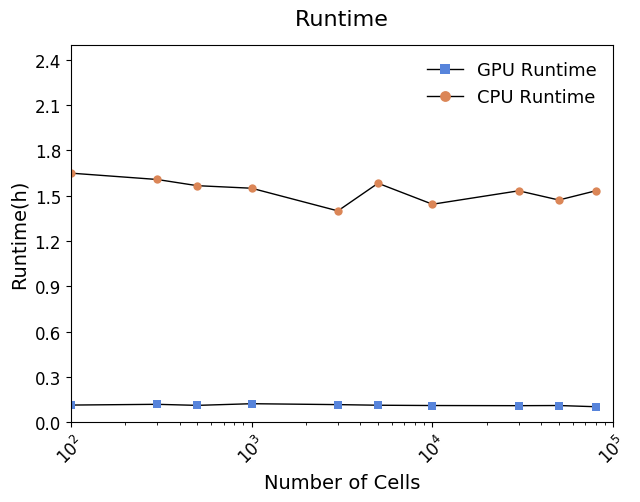
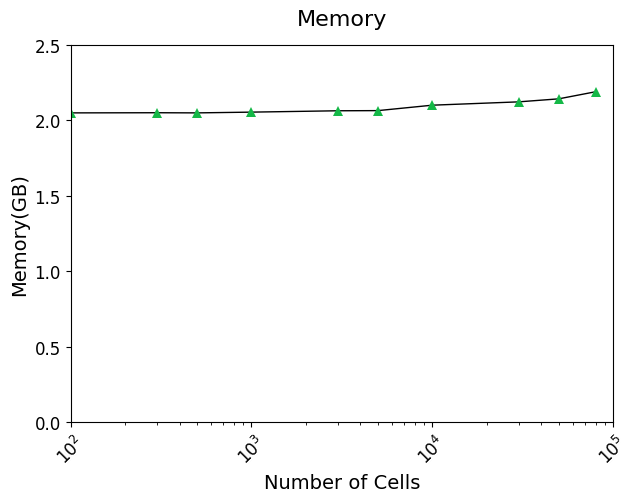
Tutorial
Tutorial Forebrain Run SCALE on dense matrix Forebrain dataset (k=8, 2088 cells)

