this is a working document
Py-oopsi: the python implementation of the fast-oopsi algorithm
Fast-oopsi was developed by joshua vogelstein in 2009, which is now widely used to extract neuron spike activities from calcium fluorescence signals. Here, we propose detailed implementation of the fast-oopsi algorithm in python programming language.
Py-oopsi requires numpy, scipy and matplotlib.
Generate Synthetic Calcium Trace
To generate synthetic calcium trace, you can
T = 2000
dt = 0.020
lam = 0.1
tau = 1.5
sigma = 0.2
# signal generator
F,C,N = oopsi.fcn_generate(T, dt=dt, lam=lam, tau=tau, sigma=sigma)where F is the Fluorescence signal with noise, C is the clean calcium trace, N is the ground truth spikes.
Reconstruct Spikes via py-oopsi
We provide demo.py to illustrate the usage of py-oopsi (as well as wiener filter, discretized binning),
# fast-oopsi,
d,Cz = oopsi.fast(F,dt=dt,iter_max=6)
# wiener filter,
d,Cw = oopsi.wiener(F,dt=dt,iter_max=100)
# descritized binning,
d,v = oopsi.discretize(F,bins=[0.75])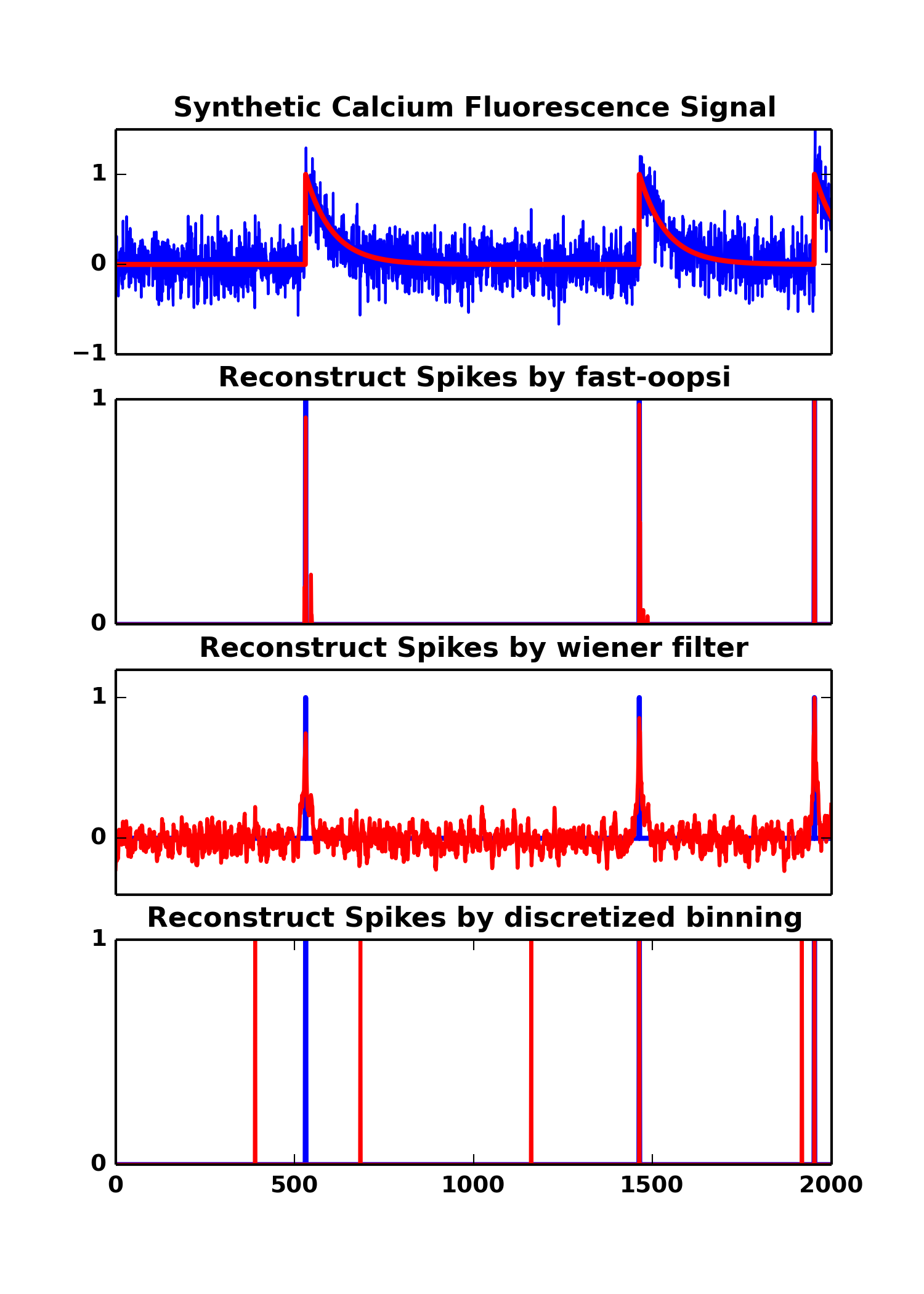
Tweak py-oopsi
py-oopsi requires
Fthe fluorescence signal, anumpy.ndarrayobject of 1-D vector;dtthe frame interval, 1/(frame rate);iter_maxmaximum number of iteration;updatetrue if the parameters are updated after each iteration.
when imaging large population of fluorescene signals of multiple neurons, for example, the connectomics challenge at kaggle.com, you need to write a subroutine to process the fluorescence trace per neuron.
Reference
- Joshua T Vogelstein, Adam M Packer, Tim A Machado, Tanya Sippy, Baktash Babadi, Rafael Yuste, Liam Paninski [Fast non-negative deconvolution for spike train inference from population calcium imaging] (http://stat.columbia.edu/~liam/research/pubs/vogelstein-fast.pdf) Journal of Neurophysiology, 104(6): 3691-3704