c2s
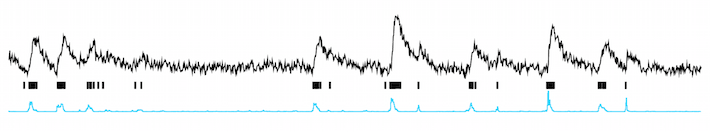
Tools for the prediction of spike trains from calcium traces.
Documentation
If you are a neuroscientist and want to reconstruct spikes from fluorescence/calcium traces or
similar signals, please see the main documentation.
If you are a developer and want to use c2s in your own Python code, please see the API documentation.
Example
Once installed, predicting spikes can be as easy as
$ c2s predict data.mat predictions.matThis package comes with a default model for predicting spikes from calcium traces, but also comes with tools for training and evaluating your own model.
Requirements
- Python >= 2.7.0
- cmt >= 0.5.0
- NumPy >= 1.6.1
- SciPy >= 0.13.0
- Cython >= 0.20.0 (optional)
- Matplotlib >= 1.4.2 (optional)
Installation
First install the Conditional Modeling Toolkit. Then run:
$ pip install git+https://github.com/lucastheis/c2s.gitYou can avoid manually installing c2s and its requirements by using Docker. A Dockerfile for c2s is provided by Jonas Rauber. This might make your life easier especially if you are planning to use Windows or Mac OS.
References
If you use our code in your research, please cite the following paper:
L. Theis, P. Berens, E. Froudarakis, J. Reimer, M. Roman-Roson, T. Baden, T. Euler, A. S. Tolias, et al.
Benchmarking spike rate inference in population calcium imaging
Neuron, 90(3), 471-482, 2016
The default model was trained on many datasets (together containing roughly 110,000 spikes) from different labs. Therefore, if you use the default model for prediction, please also cite:
J. R. Cotton, E. Froudarakis, P. Storer, P. Saggau, and A. S. Tolias
Three-dimensional mapping of microcircuit correlation structure
Frontiers in Neural Circuits, 2013
J. Akerboom et al.
Optimization of a GCaMP calcium indicator for neural activity imaging
Journal of Neuroscience, 2012
T. W. Chen et al.
Ultrasensitive fluorescent proteins for imaging neuronal activity
Nature, 2013