About
Segmentation tools based on the graph cut algorithm. You can see video to get an idea. There are two algorithms implemented. Classic 3D Graph-Cut with regular grid and Multiscale Graph-Cut for segmentation of compact objects.
please cite:
@INPROCEEDINGS{jirik2013,
author = {Jirik, M. and Lukes, V. and Svobodova, M. and Zelezny, M.},
title = {Image Segmentation in Medical Imaging via Graph-Cuts.},
year = {2013},
journal = {11th International Conference on Pattern Recognition and Image Analysis: New Information Technologies (PRIA-11-2013). Samara, Conference Proceedings },
url = {http://www.kky.zcu.cz/en/publications/JirikMmjirik_2013_ImageSegmentationin},
}Authors
- Miroslav Jirik
- Vladimir Lukes
Special requirements
See third party licenses
Resources
https://github.com/mjirik/imcut
https://github.com/amueller/gco_python
License
New BSD License, see the LICENSE file.
Install with conda (recommended)
conda install -c mjirik -c conda-forge imcut pygcoSometimes (on Linux) you will need to install pygco with pip
conda install pip
pip install pygcoInstall with pip
pip install pygco imcutSee INSTALL.md file for more information
Additional packages
conda install -c mjirik -c conda-forge seededitorqtUnderstanding the seeds
The intensity of the seed is used to train the intensity model based on the Gaussian mixture. The location of the voxel is used to set a hard constraint in the graph.
- 0 - we have no information about this voxel
- 1 - for sure, the voxel on the same location in the image data is the segmented object
- 2 - for sure, the voxel on the same location in the image data is the background
There are two more types used when the voxel is object or background but you dont want to use it for intensity model training because its intensity is not good representation.
- 3 the voxel is the object but we do not want to use it for intensity training
- 4 the voxel is the background but we do not want to use it for intensity training
The output segmentation:
- 0 - trained from
seeds==1(object) - 1 - trained from
seeds==2(background)
The only difference between object and background is in the postprocessing with the connected object filter.
It can be turned off by setting "return_only_object_with_seeds":True in segparams.
Examples
Small example
import imcut.pycut
import numpy as np
im = np.random.random([5, 5, 1])
im[:3, :3] += 1.
seeds = np.zeros([5, 5, 1], dtype=np.uint8)
seeds[:3,0] = 1 # foreground
seeds[:3,4] = 2 # background
gc = imcut.pycut.ImageGraphCut(im)
gc.set_seeds(seeds)
gc.run()
print(gc.segmentation.squeeze())[[0 0 0 1 1]
[0 0 0 1 1]
[0 0 0 1 1]
[1 1 1 1 1]
[1 1 1 1 1]]Run with CLI
Create output.mat file:
python imcut/dcmreaddata.py -i directoryWithDicomFiles --degrad 4See data:
python imcut/seed_editor_qt.py -f output.matMake graph_cut:
python imcut/pycut.py -i output.matUse is as a library:
import numpy as np
import imcut.pycut as pspc
data = np.random.rand(30, 30, 30)
data[10:20, 5:15, 3:13] += 1
data = data * 30
data = data.astype(np.int16)
igc = pspc.ImageGraphCut(data, voxelsize=[1, 1, 1])
seeds = igc.interactivity()
More complex example without interactivity
import numpy as np
import imcut.pycut as pspc
import matplotlib.pyplot as plt
# create data
data = np.random.rand(30, 30, 30)
data[10:20, 5:15, 3:13] += 1
data = data * 30
data = data.astype(np.int16)
# Make seeds
seeds = np.zeros([30,30,30])
seeds[13:17, 7:10, 5:11] = 1
seeds[0:5:, 0:10, 0:11] = 2
# Run
igc = pspc.ImageGraphCut(data, voxelsize=[1, 1, 1])
igc.set_seeds(seeds)
igc.run()
# Show results
colormap = plt.cm.get_cmap('brg')
colormap._init()
colormap._lut[:1:,3]=0
plt.imshow(data[:, :, 10], cmap='gray')
plt.contour(igc.segmentation[:, :,10], levels=[0.5])
plt.imshow(igc.seeds[:, :, 10], cmap=colormap, interpolation='none')
plt.show()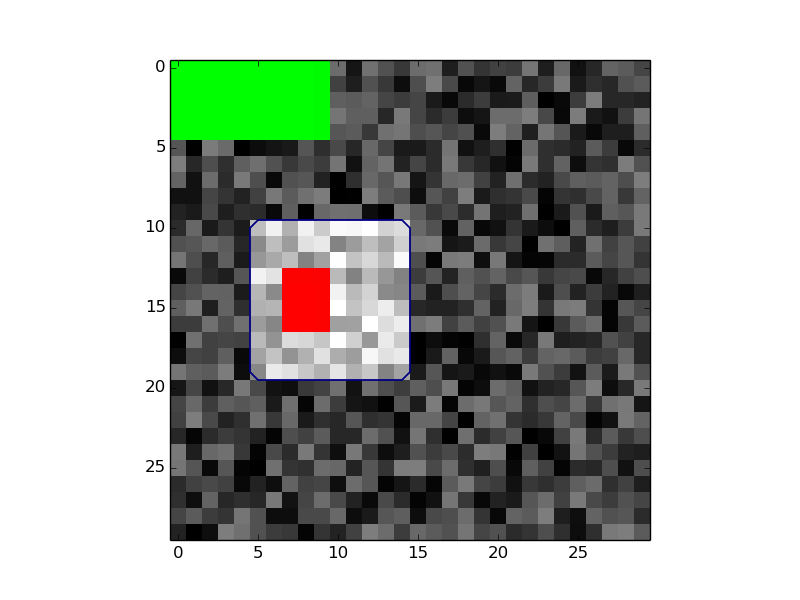
More example
- Pretrain the model to make things faster
- Use additional information about pixels
- Use custom feature vector
- See the likelihood in image and use different density functions
- Use custom density functions
Configuration
One component Gaussian model and one object
pairwise_alpha control the complexity of the object shape. Higher pairwise_alpha => more compact shape.
segparams = {
'method': 'graphcut',
"pairwise_alpha": 20,
'modelparams': {
'cvtype': 'full',
"params": {"covariance_type": "full", "n_components": 1},
},
"return_only_object_with_seeds": True,
}Gaussian mixture distribution model with extern feature vector function
segparams = {
# 'method':'graphcut',
'method': 'graphcut',
'use_boundary_penalties': False,
'boundary_dilatation_distance': 2,
'boundary_penalties_weight': 1,
'modelparams': {
'type': 'gmmsame',
'fv_type': "fv_extern",
'fv_extern': fv_function,
'adaptation': 'original_data',
},
'mdl_stored_file': False,
}mdl_stored_file: if this is set, load model from file, you can see more in function test_external_fv_with_save in pycut_test.py