
Project RACCOON
Automated construction of atomistic and coarse-grained models in the PDB format for linear polymer peptide conjugates.
Table of Contents
General Purpose & Scope
Project RACCOON (Rapid Automated Construction of Conjugates using the Optimized OPLS Input) is a Python tool designed for the generation of PDB (Protein Data Bank) files for polymer peptide conjugates, polypeptides, and polymers in a building block fashion. It allows for the easy addition of new monomers and incorporation of polypeptide and polymer sequences in text form and outputs PDB and XYZ files. The tool was specifically developed for the OPLS force field but is adaptable to any other force field. The tool's primary scope is to provide a straightforward method for creating starting structures for molecular dynamics simulations. It caters to creating linear polymer peptide conjugates with facing polypeptide strands. The tool emphasizes user-friendliness, making it accessible to chemists and physicists, not just limited to theoretical chemists with extensive background knowledge. Additionally, users can modify the code for their specific projects. Early developments started in January 2023. The software was developed at the KOMET and Besenius' research groups at the Johannes Gutenberg University Mainz.
Supported building blocks
[!NOTE] We encourage users to share their updated monomers.json file. Please submit a pull request, open an issue, or contact the developers Please refer to our contributing guidelines for further information.
| Residue Name | AA | UA | CG |
|---|---|---|---|
| PEO | :white_check_mark: | :white_check_mark: | :soon: |
| GLY | :white_check_mark: | :soon: | :soon: |
| PHE | :white_check_mark: | :white_check_mark: | :soon: |
| HIS | :white_check_mark: | :white_check_mark: | :soon: |
| ACE | :white_check_mark: | :white_check_mark: | :x: |
| NME | :soon: | :soon: | :x: |
| DUM | :white_check_mark: | ||
| CSX | :white_check_mark: | :white_check_mark: | :x: |
| LNK | :white_check_mark: | :white_check_mark: | :x: |
For non-standard amino acids CSX and LNK, please refer to the following paper by pending, 20xx. Coarse-grained models have not been implemented yet. The DUM building block is a dummy bead with an unspecified mass sometimes used in polymer modeling. Therefore, only the CG resolution is available.
How to use Project RACCOON
Installing for Standard Usage
The easiest way to use the Project RACCOON software is to install it via pip. Python >=3.11 is required, therefore it is advisable to create a conda environment
conda create -n raccoon python=3.11
conda activate raccoon
pip install project-raccoonIf you want to have access to the GROMACS test functions, you can alternatively clone the github repository and install the module via
git clone https://github.com/moritzobenauer/ProjectRaccoon.git
pip install -e .To start the command-line interface, run
project_raccoon -s {yoursequencefile}Importing new Monomers
Open a graphical software to create three-dimensional molecular models and obtain the desired monomer structure. It is important to note that only the atoms or beads of the repeating units are used and not the entire molecule. The structure must be saved in .bs format, as the position of the atoms or beads and the neighboring atoms or beads are specified here. An example alanine.bs file is provided in the examples folder. Start the user interface and navigate to Manage Monomers and Add Monomer to add the new monomer unit to your library. Please enter the name of the new residue and some properties and give every atom a unique identifier according to the force field you plan to use. For amino acid residues, it is essential to specify the C- and N-terminus of the building block.
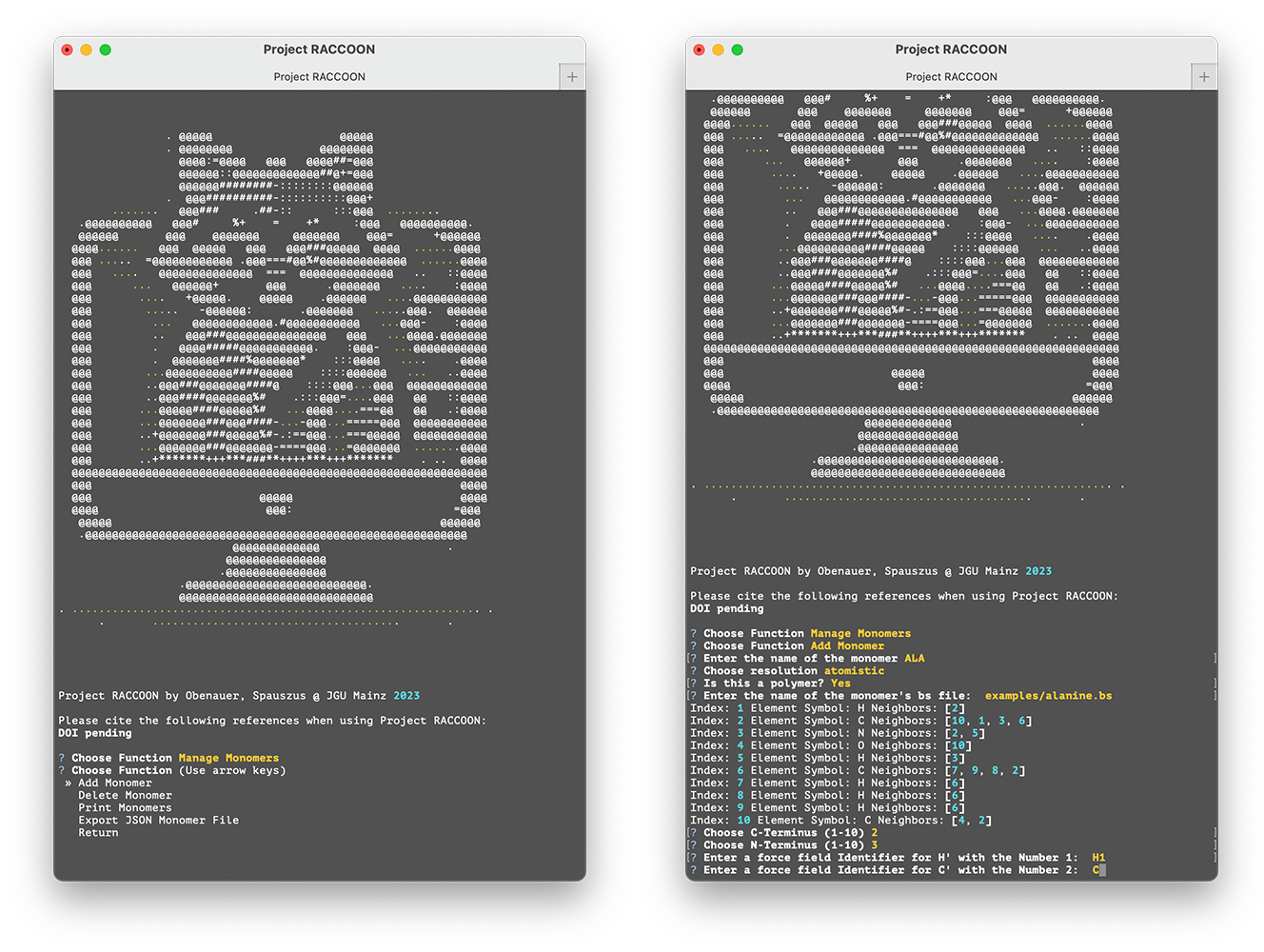
Monomer building blocks need to be given a name (no restrictions, will be treated as a string), the resolution (atomistic, united_atom, coarse_grained), and the property polymer which can be either True or False. The software will read the element type and bonded neighbors from the .bs file. The user has to set the forcefield identifiers for every single atom/bead and specifiy the C- and N-termini of the building block. After the completion of these steps, the newly added monomer is added to the available monomers list.
Creating PDB Files from Sequence
The basis for creating a PDB file is a seq.txt file containing the block sequence. The monomers are specified line by line with their name and resolution (this may be necessary if the same block is present in the library file with a different resolution). In addition, whether it is an inverted building block (this occurs with C2 symmetrical polymer peptide conjugates) and the number of repeating units must be specified. In this way, classical polymers with a fixed number of repeating units of a monomer can also be represented quickly.
HIS:AA:0:10
PHE:AA:0:10
HIS:AA:0:10The example shown here corresponds to a polypeptide in which ten histidine units are followed by ten phenylalanines and then ten histidines again. All monomers are shown in atomistic resolution and are not inverted.
| Residue Name | Resolution | Inverted | Repeats |
|---|---|---|---|
| String | {AA, UA, CG} | {0,1} | Integer |
To create the PDB file, select the first item that appears in the selection. The PDB file is then generated automatically according to the selected parameters. Creating an XYZ file from the PDB file may be desired for visualization or other purposes. This XYZ file no longer contains any binding information. For this purpose, the option Convert PDB to XYZ file can be selected in the menu. A corresponding XYZ file with the same file name as the PDB file is created.
| Parameter | Default | Description |
|---|---|---|
| -s | seq.txt | Filename with extension for the sequence. |
| -o | out.pdb | Filename without extension for the resulting pdb file. |
| -m | if None | An internal json file is used |
| -e | False | Write all explicit bonds. It can be useful for nonstandard residues. |
| -r | True | Remove duplicate bonds. Bond a --> b is equivalent to b --> a. |
You can get more control over the geometry generation by importing Project RACCOON in a notebook environment and calling the generate_file() function. The trr parameter represents a minimal threshold between two atoms/beads when applying the self-avoiding random walk geometry generation. For longer chains, it is recommended to lower this value to speed up the geometry calculation. Alternatively, the cartesian shift between two monomeric units can be adjusted by setting upper and lower bounds for the random shift. E.g., if you want to produce an elongated chain along the z-direction set shift_cartesian to [-1, 1, -1, 1, -1, 0, 2]. Lastly, setting the damping_factor to lower values can decrease the distances between monomers for long chains.
rc.generate_file(monomers, seq, False, "out.pdb", trr=1, shift_cartesian=[-1, 1, -1, 1, -1, 1, 1], damping_factor=0.5)Check PDB Files
The successful creation of the PDB file can then be checked. The Check PDB file option is selected for this purpose. If the PDB file appears in tabular form in the terminal, it is correct and will be read correctly by all standard programs. Individual atoms or beads may be arranged too close to each other. In the worst case, this can lead to problems with energy minimization or simulations. To check that no two atoms or beads are too close to each other, the Check Minimal Distance option can be selected. The output contains the smallest distance between two atoms or beads.
Examples and Testing
The software was developed specifically for the telechelic polymer peptide conjugates, according to Otter et al., 2018. An example is the peptide sequence FHFHFXG-PEO(N)-GXFHFHF (with X: 6-aminohexanoic acid). A corresponding seq.txt file can be found in the examples folder. A graphical representation of the PDB file FHFHFXG_PEO_XGFHFHF.pdb (also in the examples folder) is shown in the image below.

Please consider that the geometry shown here is not necessarily the geometry you will create when running this example. The geometry generation is partly random, yielding different structures with every run.
After a brief energy minimization (performed with e.g. GROMACS and the OPLS force field), the physically meaningful structure is obtained. The PDB file created with Project RACCOON is an excellent starting point for such molecular dynamics simulations. It is important to recognize here that different building blocks with atomistic and united atom resolution have been combined with each other.
Unit Testing
Over 90% of the code presented here is covered by unit tests. These can be run with the following command:
python -m unittest -vIf the resulting output creates no errors and prints OK, the software was correctly installed, and all features can be used. We are continuously working on improving the code coverage of the unit tests. Feel free to contribute by contacting the authors or forking the project.
GROMACS Testing
To ensure seamless integration with GROMACS, we prepared a GROMACS testing script, which can be found as gmx/automated_testing.sh. This script creates a polypeptide test structure and runs the gmx pdb2gmx, gmx grompp, and gmx mdrun commands to ensure that the created pdb files can be read by GROMACS. By also running an energy minimization, we guarantee the creation of geometries that do not lead to infinite forces. The folder contains the necessary GROMACS input files and the FHFHF test sequence as a text file.
Limitations
[!WARNING]
- The generated output is only a starting structure for molecular dynamics simulations.
- Users must ensure atom/bead naming consistency for matching force field data.

