Generative GaitNet (Simple Version)
The updated version is available in https://github.com/namjohn10/BidirectionalGaitNet.
Abstract
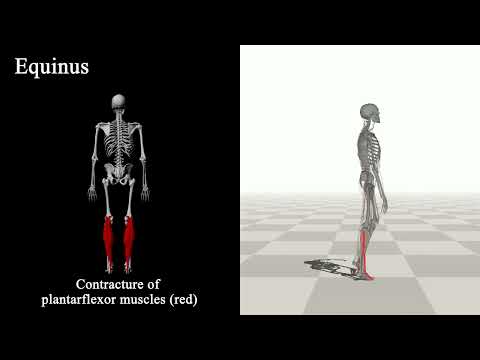
Understanding the relation between anatomy and gait is key to successful predictive gait simulation. In this paper, we present Generative GaitNet, which is a novel network architecture based on deep reinforcement learning for controlling a comprehensive, full body, musculoskeletal model with 304 Hill-type musculotendons. The Generative GaitNet is a pre-trained, integrated system of artificial neural networks learned in a 618-dimensional continuous domain of anatomy conditions (e.g., mass distribution, body proportion, bone deformity, and muscle deficits) and gait conditions (e.g., stride and cadence). The pre-trained GaitNet takes anatomy and gait conditions as input and generates a series of gait cycles appropriate to the conditions through physics-based simulation. We will demonstrate the efficacy and expressive power of Generative GaitNet to generate a variety of healthy and pathological human gaits in real-time physics-based simulation.
Video (Youtube)
Publications
https://mrl.snu.ac.kr/research/ProjectGaitNet/paper.pdf
Jungnam Park, Sehee Min, Phil Sik Chang, Jaedong Lee, Moon Seok Park, and Jehee Lee Generative GaitNet, SIGGRAPH 2022 Conference Proceedings.
Installation
We checked code works in Python 3.6, ray(rllib) 1.8.0 and Cluster Server (64 CPUs (128 threads) and 1 GPU (RTX 3090) per node)
Install Library Automatically
cd {downloaded folder}/
./install.shCompile
cd {downloaded folder}/
./pc_build.sh
cd build
make -j32Rendering
To run without policy
cd {downloaded folder}/build
./imgui_render/imgui_render ../data/metadata.txtor the trained policy.
cd {downloaded folder}/build
./imgui_render/imgui_render {network_path}To run with trained 4 policies (to lower body),
cd {downloaded folder}/build
./imgui_render/imgui_render ../data/trained_nn/Skeleton ../data/trained_nn/Ankle ../data/trained_nn/Hip ../data/trained_nn/MergeAnd check the edge connection file, ./data/cascading_map.txt (The right number is descendant policy in the graphs)
1 0
2 0
3 0
3 1
3 2Learning
Parameter Setting
Set adjustable parameters in {downloaded folder}/data/metadata.txt. The group flag groups muscles containing the given name.
To set muscle length,
muscle_param
group(or not use) {Left Muscle1 Name} {min ratio} {max ratio}
group(or not use) {Right Muscle1 Name} {min ratio} {max ratio}
group(or not use) {Left Muscle2 Name} {min ratio} {max ratio}
group(or not use) {Right Muscle2 Name} {min ratio} {max ratio}
...
muscle_endTo set muscle force,
muscle_param
group(or not use) {Left Muscle1 Name} {min ratio} {max ratio}
group(or not use) {Right Muscle1 Name} {min ratio} {max ratio}
group(or not use) {Left Muscle2 Name} {min ratio} {max ratio}
group(or not use) {Right Muscle2 Name} {min ratio} {max ratio}
...
muscle_endTo set body proportion force,
muscle_param
global {min ratio} {max ratio}
Head {min ratio} {max ratio}
{Left Body Name} {min ratio} {max ratio}
{Right Body Name} {min ratio} {max ratio}
...
muscle_endTraining
Training is executed based on the metadata setting.
Training a single policy (Cluster setting)
cd {downloaded folder}/python
python3 ray_train.py --config=ppo_medium_node Cascading and Subsumption learning
cd {downloaded folder}/python
python3 ray_train.py --config=ppo_medium_node --cascading_nn={previous network paths}