RamanNet
A generalized neural network architecture for Raman Spectrum Analysis
This repository contains the original implementation of "RamanNet : A generalized neural network architecture for Raman Spectrum Analysis" in Tensorflow
Introduction
Raman spectroscopy provides a vibrational profile of the molecules and thus can be used to uniquely identify different kind of materials. This sort of fingerprinting molecules has thus led to widespread application of Raman spectrum in various fields like medical dignostics, forensics, mineralogy, bacteriology and virology etc. Despite the recent rise in Raman spectra data volume, there has not been any significant effort in developing generalized machine learning methods for Raman spectra analysis. We examine, experiment and evaluate existing methods and conjecture that neither current sequential models nor traditional machine learning models are satisfactorily sufficient to analyze Raman spectra. Both has their perks and pitfalls, therefore we attempt to mix the best of both worlds and propose a novel network architecture RamanNet. RamanNet is immune to invariance property in CNN and at the same time better than traditional machine learning models for the inclusion of sparse connectivity. Our experiments on 4 public datasets demonstrate superior performance over the much complex state-of-the-art methods and thus RamanNet has the potential to become the defacto standard in Raman spectra data analysis
Model Overview
The proposed model breaks a Raman spectrum into overlapping segments and processes them using shifted MLP layers. In this way the weights are not shared and we only need to process a small portion at a time.
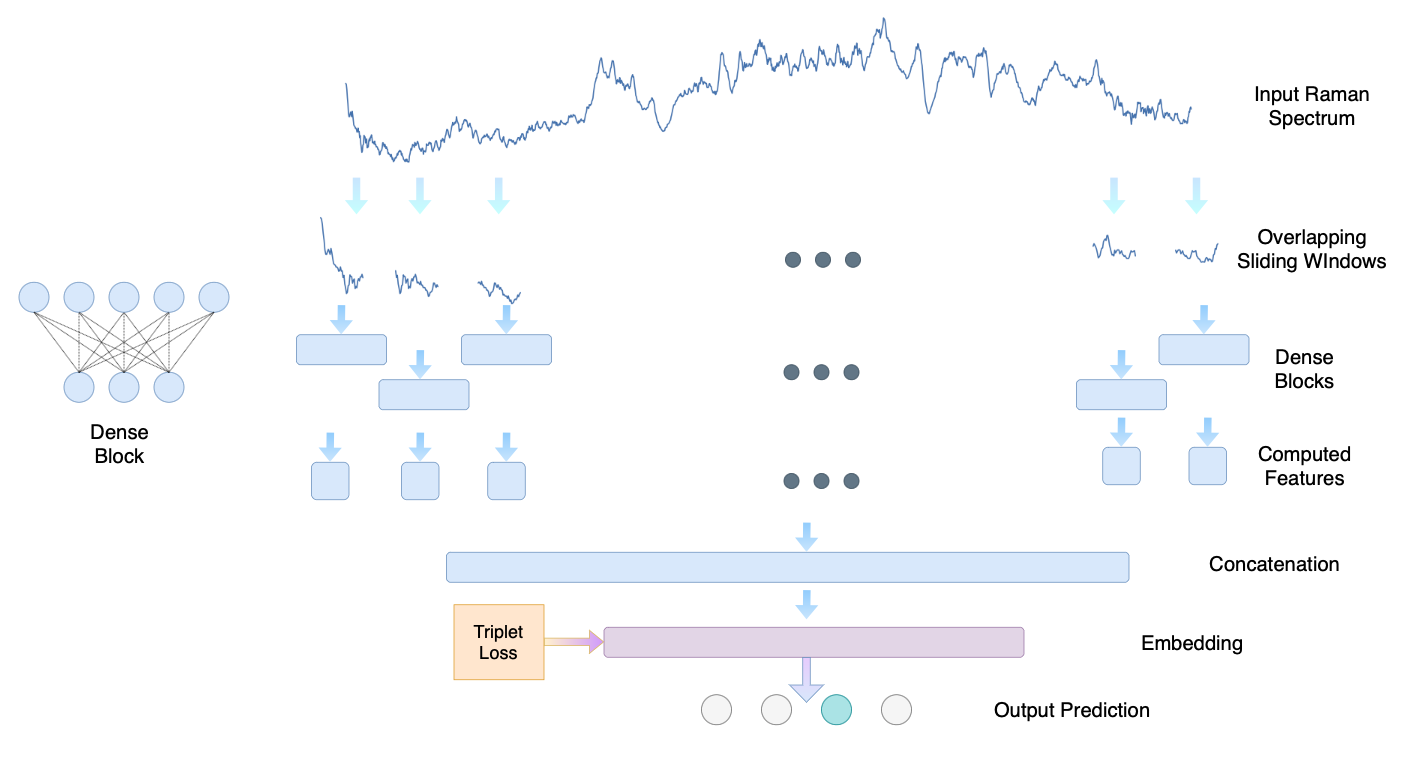
Codes
We provide the codes for Raman spectrum processing along with RamanNet model and training. Our codes are based on tensorflow and numpy. The codes can be found in the /codes directory.
- data_processing.py : contains helper function for data procssing
- RamanNet_model.py : contains the model definition
- train_model.py : contains the training script
Tutorial
You can use RamanNet on your own Raman spectrum dataset.
-
Collect the Raman spectrum, label them and split them into folds. Our model is designed to be a general one and we don't have any particular restrictions on the type of spectra.
-
Compute the overlapping segments of the spectrum using our provided segment_spectrum_batch function. This function takes an array of the Raman spectra, and two (optional) parameters w (length of window. Defaults to 50) and dw (step size. Defaults to 25).
from codes.data_processing import segment_spectrum_batch
windowed_spectra = segment_spectrum_batch(input_raman_spectra_array, w = 50, dw = 25)-
Do the same for both training and validation spectra. Also convert the labels to one-hot-encoding.
-
Create an instance of RamanNet model and train it using like standard keras models.
from codes.RamanNet_model import RamanNet
mdl = RamanNet(w_len, n_windows, n_classes)- Alternatively, you may skip all the previous steps and use our
train_modelfunction, which combines all these steps. All you need is to provide the training and validation of raman spectrum arrays and one-hot encoding of labels, window length and step size, number of epochs and path to save the model. This function will convert the spectrum to windowed spectrums, create an instance of the model, the model will be trained and saved in the model_path
from codes.train_model import train_model
trained_model, training_history = train_model(X_train, Y_train_onehot, X_val, Y_val_onehot, w_len, dw, epochs, model_path)- Validate on your test data or deploy.
Web server
We have prepared a web server to detect Bacterial Infection using RamanNet
Citation Request
If you use RamanNet in your project, please cite the following paper
@article{ibtehaz2022ramannet,
title={RamanNet: a generalized neural network architecture for Raman spectrum analysis},
author={Ibtehaz, Nabil and Chowdhury, Muhammad EH and Khandakar, Amith and Zughaier, Susu M and Kiranyaz, Serkan and Rahman, M Sohel},
journal={arXiv preprint arXiv:2201.09737},
year={2022}
}