Chronologer
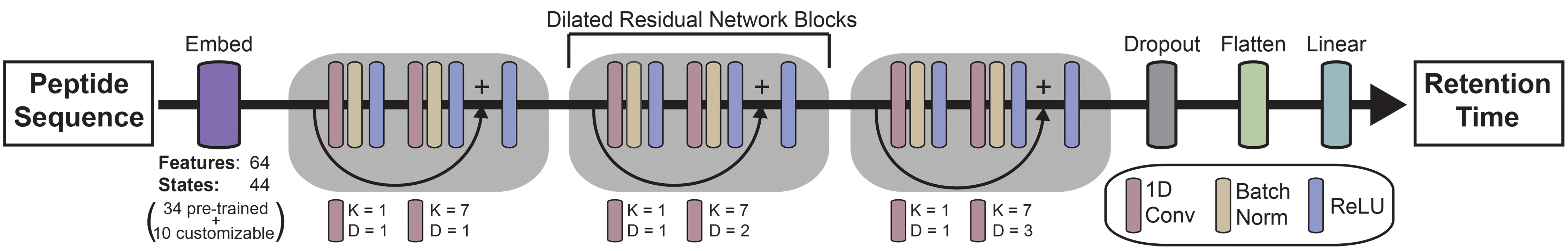 Chronologer is a deep learning model for highly accurate prediction of peptide C18 retention times (reported in % ACN). Chronologer was trained on a new large harmonized database of >2.6 million retention time observations (2.25M unique peptides) constructed from 11 community datasets and natively supports prediction of 17 different modification types. With only a few observations of a new modification type (>10 peptides), Chronologer can be easily re-trained to predict up to 10 user supplied modifications.
Chronologer is a deep learning model for highly accurate prediction of peptide C18 retention times (reported in % ACN). Chronologer was trained on a new large harmonized database of >2.6 million retention time observations (2.25M unique peptides) constructed from 11 community datasets and natively supports prediction of 17 different modification types. With only a few observations of a new modification type (>10 peptides), Chronologer can be easily re-trained to predict up to 10 user supplied modifications.
Installation
Option 1
Clone the github repository into the desired directory.
| Chronologer has been verified to work under the following environment: | Package | Version |
|---|---|---|
| Python | 3.10.6 | |
| PyTorch | 1.21.1 | |
| Numpy | 1.23.2 | |
| Pandas | 1.4.3 | |
| Scipy | 1.9.1 | |
| Pyteomics | 4.5.5 |
Option 2
It is highly encouraged to use virtual environments to isolate the package.
From the command line this can be done like so:
python3 -m venv venv # creates an environment called venv
source venv/bin/activate # activates itThe package can then be installed from github directly like so:
python -m pip install "chronologer @ git+https://github.com/searlelab/chronologer.git"And we can check if it installed correctly using the chronologer command.
chronologer_predict --helpUsage
Chronologer can predict C18 retention coefficients for peptides between 6-50 amino acids in length and any of the supported modifications:
| Modification | Mass | Residues |
|---|---|---|
| Carbamidomethyl | +57.021464 | C |
| Oxidation | +15.99491 | M |
| Phosphorylation | +79.966331 | S, T, Y |
| Acetylation | +42.010565 | K, N-term |
| Succinylation | +101.023869 | K |
| Ubiquitylation | +114.042927 | K |
| Mono-methylation | +14.015650 | K, R |
| Di-methylation | +28.031300 | K, R |
| Tri-methylation | +42.046950 | K |
| TMT0 | +224.152478 | K, N-term |
| TMT10 | +229.162932 | K, N-term |
| Pyroglutamate | -18.010565 | N-term E |
| Pyroglutamate | -17.026549 | N-term Q |
| Cyclized S-CAM-Cys | +39.994915 | N-term C |
Chronologer uses EncyclopeDIA-style formatting for modified peptides where modification mass shifts after included in [] after the modified residue. E.g.
| PeptideModSeq | Modification |
|---|---|
| ALSVLGC[+57.021464]GHTSSTK | Carbamidomethyl at C7 |
| ASPGTPLSPGS[+79.966331]LR | Phospho at S11 |
| [42.010565]KGSPTPGFSTR | N-terminal acetylation |
The retention time prediction script (Predict_RT.py) takes as its input a tab-separated value (TSV) file that must include a "PeptideModSeq" column with EncyclopeDIA-style sequences (for pre-trained modifications, 2 decimal points of mass resolution is sufficient), and will return a new TSV with a "Pred_HI" column containing the Chronologer predictions. Any additional columns provided in the input TSV will be preserved in the output, with rows containing Chronologer-incompatible peptides removed.
Chronologer can be re-trained to support additional modifications (mininum 10 peptides). Re-training first requires alignment of user retention times into Hydrophobic Index (HI) space (i.e. the % ACN in 0.1% FA that a peptide will elute). As long as user source data contains some Chronologer-predictable peptides (preferably >100), this can be easily done using the provided Align_RT_to_Hydrophobic_Index.py script (use -h flag for detailed instructions). To train Chronologer for new modifications, the modification masses must first be defined in the user_modifications.txt file, which is a TSV file with 3 columns: index, residue, and mass. Do not alter the index column, which is used for internal purposes. User provided masses must perfectly match the values listed in the PeptideModSeq strings. Modifications can be disabled by commenting out the line (i.e. adding # to the beginning of the line). Detailed instructions for training Chronologer can be found by running "python Train_Chronologer.py -h".
Demo
A small test file (demo_peptides.txt) with 100 "crap peptides" is included to test a working environment. From the command line, retention times can be predicted by running
python Predict_RT.py demo_peptides.txt demo_rt.txt
where "demo_rt.txt" can be replaced with the output file name of your choice