mubind


Documentation and tutorials
Please refer to the main documentation and tutorials at
https://mubind.readthedocs.io/en/latest/tutorials.html
Model highlights
- MuBind is a deep learning model that can learn DNA-sequence features predictive of cell transitions in single-cell genomics data, using graph representations and sequence-activity across cells. The codebase is written in PyTorch.
- This package works with single-cell genomics data, scATAC-seq, etc. We have also tested it on bulk in vitro samples (HT-SELEX). See documentation for examples.
- Complemented with velocity-driven graph representations we learn sequence-to-activity transcriptional regulators linked with developmental processes. These predictions are biologically confirmed in several systems, and reinforced through chromatin accessibility and orthogonal gene expression data across pseudotemporal order. Refer to bioRxiv for more details.
Workflow and model architecture
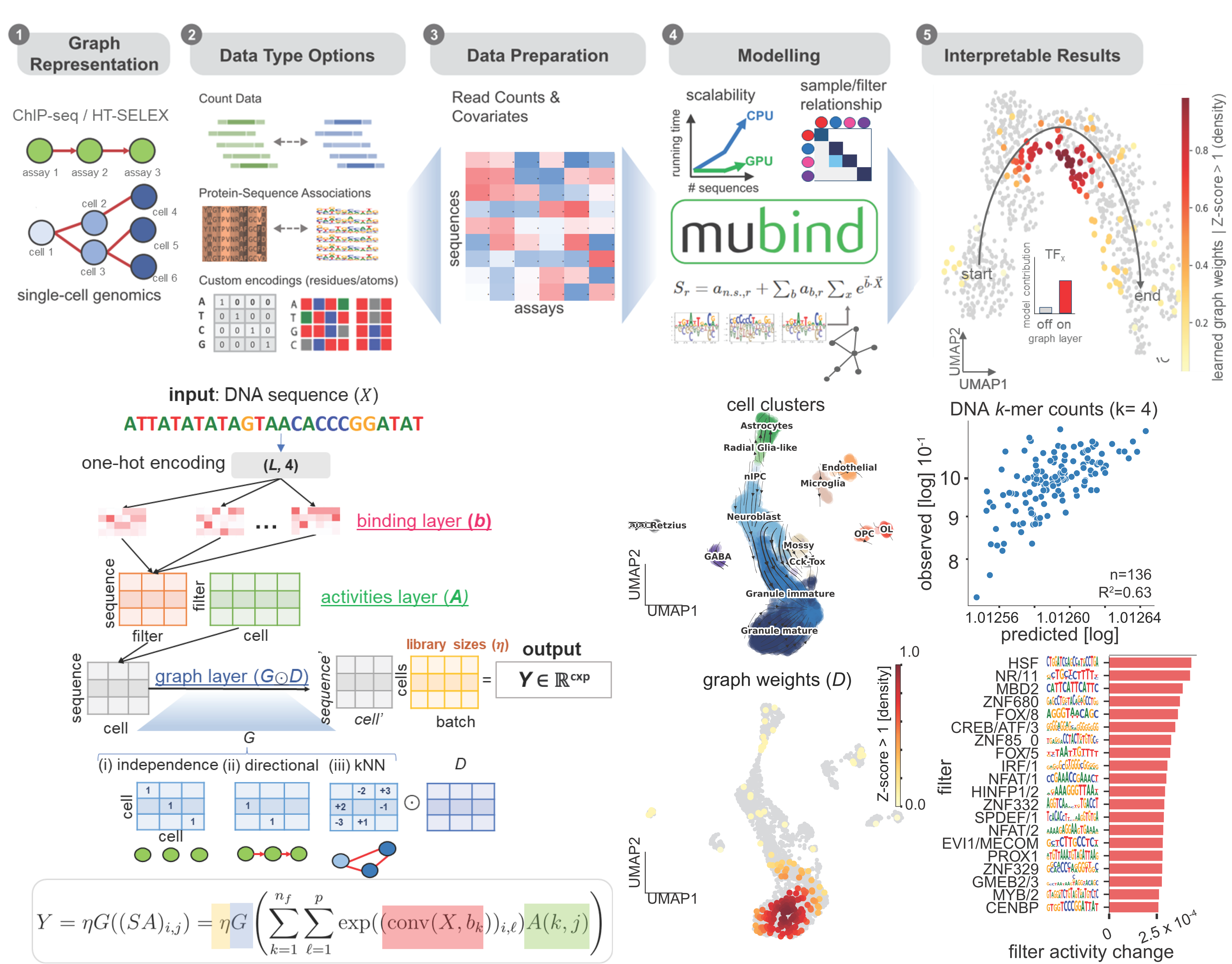
Other specifications
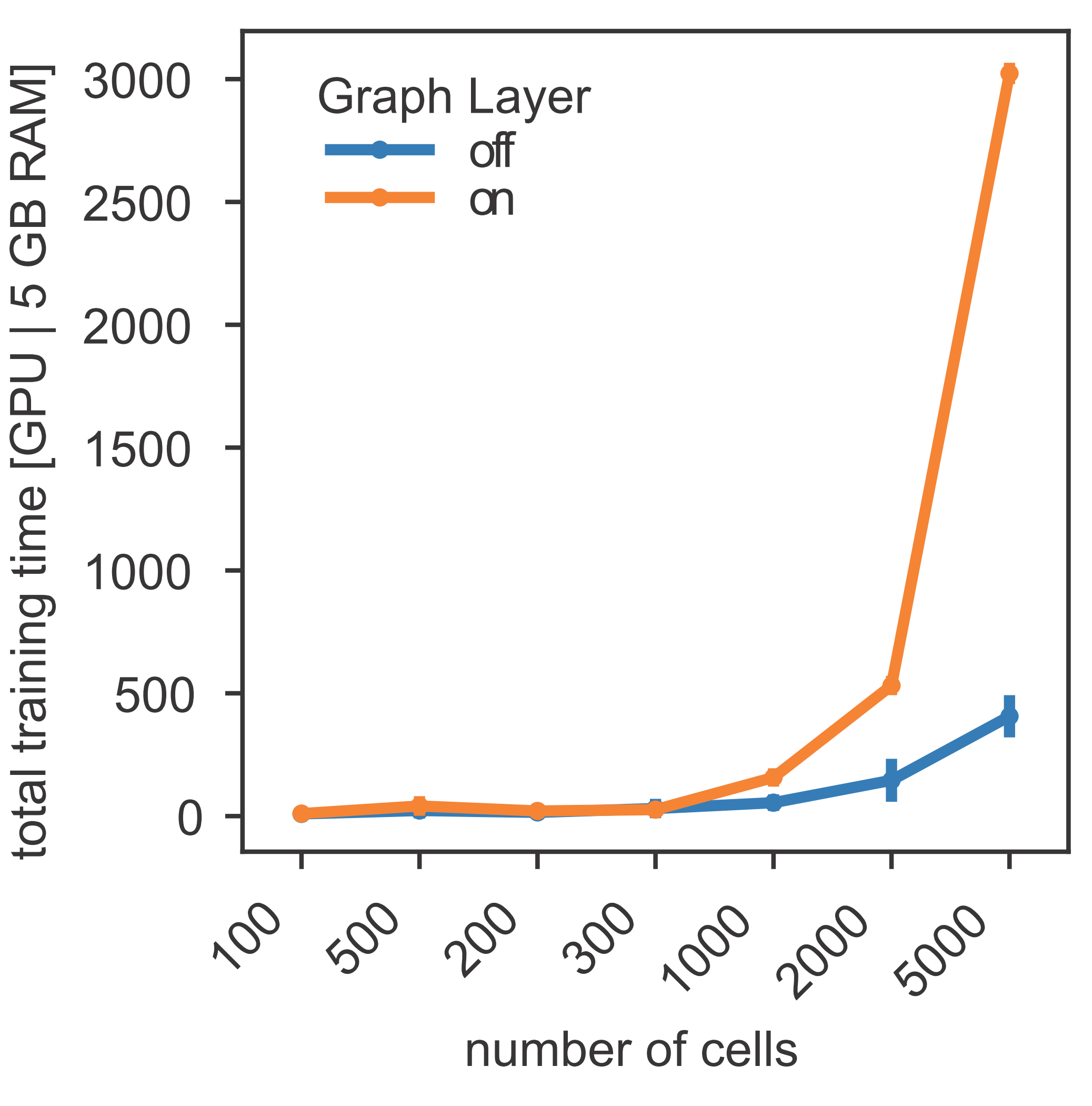
- Number of cells: The scalability of this method has been tested on single-cell datasets between 1,000 and 100,000 cells.
- Number of peaks: We have tested three-times the number of features (peaks, promoters) selected randomly and with EpiScanpy's variability score. In our experience, highest testing performances are obtained when using random features. all features requires calibration of batch sizes and total GPU memory.
- Running time: Using a Graph Layer and PWMs in the Binding Layer, the running time with one GPU is about 50 min (5,000 cells, 15,000 features). For additional memory and scaling tips, please refer to the documentation.
Installation
There are several alternative options to install mubind:
pip
- Install the latest release of
mubindfromPyPI <https://pypi.org/project/mubind/>_:
pip install mubind- Install the latest development version:
pip install git+https://github.com/theislab/mubind.git@mainRelease notes
See the changelog.
Preprint
If mubind is useful for your research, please consider citing as:
Ibarra I.L., Schneeberger J., Erdogan E., Redl L., Martens L., Klein D., Aliee H., and Theis F.J. Learning sequence-based regulatory dynamics in single-cell genomics bioRxiv 2024.08.07.605876 (2024) doi:10.1101/2024.08.07.605876.
Funding acknowledgments.
Issues
If you found a bug, please open an Issue.
Project template created using scverse cookie template