-
Hi, @tangerzhang
I have finished my contig-level assembly using hifiasm with hifi reads. Because Purge-dups was contained in hifiasm, so I directly using Purge-dups contigs. Following your tutoria…
-
Hi,
Thanks for sharing this amazing tool.
I am building the assemblies according to the homepage pipeline, and the bwa sampe is quite time-consuming,
so, at the same time, I tried to use t…
-
Here a resume of the size of my genome after each step
| assembly | primary size (bp) | haplotig size (bp) | total (bp)
| --- | --- | -- | -- |
| falcon unzip | 879494072 | 161125037 | 104061…
-
Hi, is ALLHiC compatible with Omni-C libraries? Thanks, André
-
Hi,
We have one population consisted of 192 F3 lines resulting from a cross between two plants (Lab x QLD).
Additionally, we created 2 assemblies out of the above two plants and grouped them and gro…
-
Dear Xingtan,
Do you have any plan to update multiple restriction enzyme functions?
I love you.
Won
-
Hi, @tanghaibao and @tangerzhang ,
my scaffolds have many misjoins, i konw you have much experience in processing HiC data. is there any suggestions to perform error correct before running Allhic?…
-
Hi, I have got a problem when I was running the protocol. I have finished generating the sam file from fq1 and fq2 file, The generated sam file is about 100G, but when I was running the PreprocessSAMs…
-
Questions are as follows:
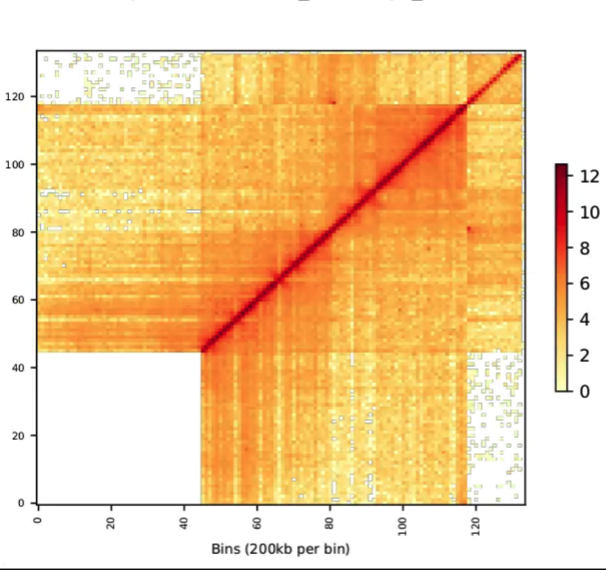
how to solve this problem? thanks.
-
I run this command as follow and the `partition.pl` generate empty file with just `chr01` directory.
```
draft_asm=litchi.contigs.fasta
partition.pl -g Allele.gene.table -r $draft_asm -b sample.cle…