-
Hi,
Sorry to bother with this request.
I have my own summary statistics that I am using as JSON file for a region and am using your server API LDLZ2 for LD information.
What I really would like to…
-
As captured by Sentry:
UKBB PheWAS plots encounter an error when viewing the following region:
https://portaldev.sph.umich.edu/ukbb/v1/statistic/phewas/?filter=variant%20eq%20%276%3A26093141_G%2FA…
-
Stephanie Gogarten of UW has a few repos worth perusing, though she uses R and not Python: https://github.com/UW-GAC/analysis_pipeline shares similar goals to this repository but for TOPMed data; http…
-
Hi,
We have been using Exac constraint API (http://exac.broadinstitute.org/api/constraint) as our data source for Locuszoom plot. But, that link hasn't been working and gives us 405 error and CORS…
-
Hi,
Thanks a lot for your contributions to the Pheweb project. It's amazing to see the results displayed on a website.
I am quite new to web development. I am exploring to deploy the website buil…
-
Hi,
I am trying to plot my own fine-mapping results to the LocusZoom plot. However, sometimes the fine-mapped region is omitted in the plot, and sometimes it shows fine.
For example, in one regi…
-
For a plot displaying a single variant which affects multiple genes and multiple tissues, we want to give the user the ability to choose to highlight results for a single gene:
- When the user cli…
-
Hi, guys:
It is amazing that IGV could also show GWAS data. But I found a few issues. It would be great if someone is interested in addressing them:
I could not move thee top “Gene” panel to t…
-
To reproduce:
1. Hover over a tooltip.
2. ctrl-tab away from the tab.
3. come back, and it stays open until I close it or re-mouse-out its point.
On , clicking a point usually makes it exclusive…
-
I'm using pyGenomeTracks to plot genes, and get this output:
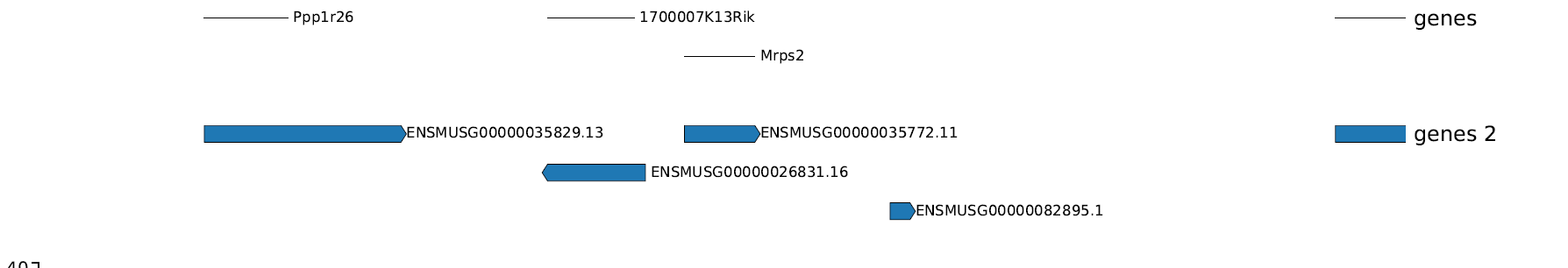
the upper track is from be…